Genetic Geometry Takes Shape
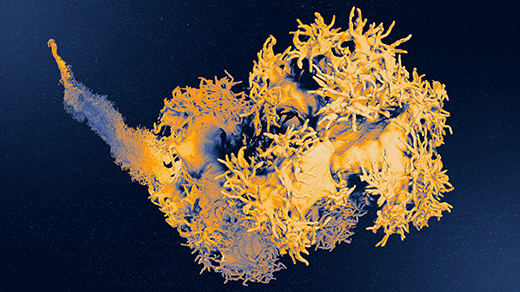
The nuclei from a half-million human cells could all fit inside a single poppy seed. Yet within each and every nucleus resides genomic machinery that is incredibly vast, at least from a molecular point of view. It has billions of parts, many used to activate and silence genes — an arrangement that allows individual cells to specialize as brain cells, heart cells and some 200 other different cell types. What’s more, each cell’s genome is atwitter with millions of mobile pieces that swarm throughout the nucleus and latch on here and there to tweak the genetic program. Every so often, the genomic machine replicates itself.
At the heart of the human genome’s Lilliputian machinery is the two meters’ worth of DNA that it takes to embody a person’s 3 billion genetic letters, or nucleotides. Stretch out all of the genomes in all of your body’s trillions of cells, says Tom Misteli, the head of the cell biology of genomes group at the National Cancer Institute in Bethesda, Md., and it would make 50 round trips to the sun. Since 1953, when James Watson and Francis Crick revealed the structure of DNA, researchers have made spectacular progress in spelling out these genetic letters. But this information-storage view reveals almost nothing about what makes specific genes turn on or off at different times, in different tissue types, at different moments in a person’s day or life.
To figure out these processes, we must understand how those genetic letters collectively spiral about, coil, pinch off into loops, aggregate into domains and globules, and otherwise assume a nucleus-wide architecture. “The beauty of DNA made people forget about the genome’s larger-scale structure,” said Job Dekker, a molecular biologist at the University of Massachusetts Medical School in Worcester who has built some of the most consequential tools for unveiling genomic geometry. “Now we are going back to studying the structure of the genome because we realize that the three-dimensional architecture of DNA will tell us how cells actually use the information. Everything in the genome only makes sense in 3-D.”
Genome archaeologists like Dekker have invented and deployed molecular excavation techniques for uncovering the genome’s architecture with the hope of finally discerning how all of that structure helps to orchestrate life on Earth. For the past decade or so, they have been exposing a nested hierarchy of structural motifs in genomes that are every bit as elemental to the identity and activity of each cell as the double helix.
A Better Genetic Microscope
A close investigation of the genomic machine has been a long time in coming. The early British microscopist Robert Hooke coined the word cell as a result of his mid-17th-century observations of a thin section of cork. The small compartments he saw reminded him of monks’ living quarters — their cells. By 1710, Antonie van Leeuwenhoek had spied tiny compartments within cells, though it was Robert Brown, of Brownian motion fame, who coined the word nucleus to describe these compartments in the early 1830s. A half-century later, in 1888, the German anatomist Heinrich Wilhelm Gottfried von Waldeyer-Hartz peered through his microscope and decided to use the word chromosome — meaning “color body” — for the tiny, dye-absorbing threads that he and others could see inside nuclei with the best microscopes of their day.
During the 20th century, biologists found that the DNA in chromosomes, rather than their protein components, is the molecular incarnation of genetic information. The sum total of the DNA contained in the 23 pairs of chromosomes is the genome. But how these chromosomes fit together largely remained a mystery.
Then in the early 1990s, Katherine Cullen and a team at Vanderbilt University developed a method to artificially fuse pieces of DNA that are nearby in the nucleus — a seminal feat that made it possible to analyze the ultrafolded structure of DNA merely by reading the DNA sequence. This approach has been improved over the years. One of its latest iterations, called Hi-C, makes it possible to map the folding of entire genomes.
The first step in a Hi-C experiment is to treat a sample of millions of cells with formaldehyde, which has the chemical effect of cross-linking strands of DNA wherever two strands happen to be close together. Those two nearby bits might be some distance away along the same chromosome that has bent back onto itself, or they may be on separate but adjacent chromosomes.
Next, researchers mince the genomes, harvest the millions of cross-linked snippets, and sequence the DNA of each snippet. The sequenced snippets are like close-up photos of the DNA-DNA contacts in the 3-D genome. Researchers map these snippets onto existing genome-wide sequence data to create a listing of the genome’s contact points. The results of this matching exercise are astoundingly data-rich maps — they look like quilts of nested, color-coded squares of different sizes — that specify the likelihood of any two segments of a chromosome (or even two segments of an entire genome) to be physically close to one another in the nucleus.
So far, most Hi-C data depict an average contact map using contact hits pooled from all of the cells in the sample. But researchers have begun to push the technique so that they can harvest the data from single cells. The emerging capability could lead to the most accurate 3-D renderings yet of chromosomes and genomes inside nuclei.
In addition, Erez Lieberman Aiden, the director of the Baylor College of Medicine Center for Genome Architecture, and his colleagues have recently cataloged DNA-DNA contacts in intact nuclei, rather than in DNA that previously had to be extracted from nuclei, a step that adds uncertainty to the data. The higher-resolution contact maps enable the researchers to discern genomic structural features on the scale of 1,000 genetic letters — a resolution about 1,000 times finer than before. It is like looking right under the hood of a car instead of squinting at the engine from a few blocks away. The researchers published their views of nine cell types, including cancer cells in both humans and mice, in the December 18, 2014, issue of Cell.
The Power of Loops
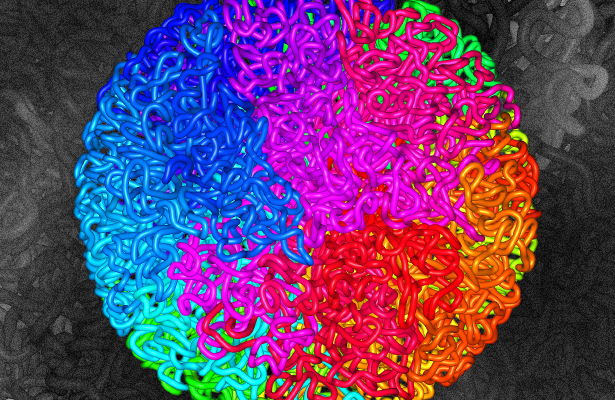
Using sophisticated algorithms to analyze the hundreds of millions — in some cases, billions — of contact points in these cells, Aiden and his colleagues could see that these genomes pinch off into some 10,000 loops. Cell biologists have known about genomic loops for decades, but were not previously able to examine them with the level of molecular resolution and detail that is possible now. These loops, whose fluid shapes Dekker likens to “snakes all curled up,” reveal previously unseen ways that the genome’s large-scale architecture might influence how specific genes turn on and off, said Miriam Huntley, a doctoral student at Harvard University and a co-author of the Cell article.
In the different cell types, the loops begin and end at different specific chromosomal locations, so each cell line’s genome appears to have a unique population of loops. And that differentiation could provide a structural basis to help explain how cells with the same overall genome nonetheless can differentiate into hundreds of different cell types. “The 3-D architecture is associated with which program the cell runs,” Aiden said.
What do these loops do? Misteli imagines them “swaying in the breeze” inside the fluid interior of the nucleus. As they approach and recede from one another, other proteins might swoop in and stabilize the transient loop structure. At that point, a particular type of protein called a transcription activator can kick-start the molecular process by which a gene gets turned on.
Misteli muses that each cell type — a liver cell or a brain cell, for example — could have a signature network of these transient loop-loop interactions. Loop structures could determine which genes get activated and which get silenced.
Yet the researchers are careful to note that they’ve only found associations between structure and function — it’s still too early to know for sure if one causes the other, and the direction in which the causal arrow points.
As they mined their data on inter-loop interactions, Aiden, Huntley and their colleagues were also able to discern a half-dozen larger structural features in the genome called subcompartments. Aiden refers to them as “spatial neighborhoods in the nucleus” — the nucleic equivalent of New York City’s midtown or Greenwich Village. And just as people gravitate toward one neighborhood or another, different stretches of chromosomes carry a kind of molecular zip code for certain subcompartments and tend to slither toward them.
These molecular zip codes are written in chromatin, the mix of DNA and protein that makes up chromosomes. Chromatin is built when DNA winds around millions of spool-like protein structures called nucleosomes. (This winding is why two meters of DNA can cram inside nuclei with diameters just one-three-hundred-thousandth as wide.)
A large cast of biomolecular players finesses different swaths of this contorted chromatin into more closed or open shapes. Roving parts of the genomic machine can better access the open sections, and so have a better chance of turning on the genes located there.
Video: How does the genome fold? Researchers use origami to explain their findings.
The increasingly detailed hierarchical picture of the genome that researchers like Dekker, Misteli, Aiden and their colleagues have been building goes something like this: Nucleotides assemble into the famous DNA double helix. The helix winds onto nucleosomes to form chromatin, which winds and winds in its turn into formations similar to what you get when you keep twisting the two ends of a string. Amid all of this, the chromatin pinches off here and there into thousands of loops. These loops, both on the same chromosome and on different ones, engage one another in subcompartments.
As researchers gradually gain more insight into the genome’s hierarchy of structures, they will get closer to figuring out how this macromolecular wonder works in all of its vastness and mechanistic detail. The National Institutes of Health has launched a five-year, $120 million program called 4D Nucleome that is sure to build momentum in the nuclear-architecture research community, and a similar initiative is being launched in Europe. The goal of the NIH program, as described on its website, is “to understand the principles behind the three-dimensional organization of the nucleus in space and time (the fourth dimension), the role nuclear organization plays in gene expression and cellular function, and how changes in the nuclear organization affect normal development as well as various diseases.”
Or, as Dekker says, “It will finally allow us to see the living genome in action, and that would ultimately tell us how it actually works.”
This article was reprinted on ScientificAmerican.com.