A Secret Flexibility Found in Life’s Blueprints

Ryan Peltier for Quanta Magazine
Introduction
The millimeter-long roundworm Caenorhabditis elegans has about 20,000 genes — and so do you. Of course, only the human in this comparison is capable of creating either a circulatory system or a sonnet, a state of affairs that made this genetic equivalence one of the most confusing insights to come out of the Human Genome Project. But there are ways of accounting for some of our complexity beyond the level of genes, and as one new study shows, they may matter far more than people have assumed.
For a long time, one thing seemed fairly solid in biologists’ minds: Each gene in the genome made one protein. The gene’s code was the recipe for one molecule that would go forth into the cell and do the work that needed doing, whether that was generating energy, disposing of waste, or any other necessary task. The idea, which dates to a 1941 paper by two geneticists who later won the Nobel Prize in medicine for their work, even has a pithy name: “one gene, one protein.”
Over the years, biologists realized that the rules weren’t quite that simple. Some genes, it turned out, were being used to make multiple products. In the process of going from gene to protein, the recipe was not always interpreted the same way. Some of the resulting proteins looked a little different from others. And sometimes those changes mattered a great deal. There is one gene, famous in certain biologists’ circles, whose two proteins do completely opposite things. One will force a cell to commit suicide, while the other will stop the process. And in one of the most extreme examples known to science, a single fruit fly gene provides the recipe for more than 38,000 different proteins.
But these are dramatic cases. It was never clear just how common it is for genes to make multiple proteins and how much those differences matter to the daily functioning of the cell. Many researchers have assumed that the proteins made by a given gene probably do not differ greatly in their duties. It’s a reasonable assumption — many small-scale tests of sibling proteins haven’t suggested that they should be wildly different.
It is still an assumption, however, and testing it is quite an endeavor. Researchers would have to take a technically tricky inventory of the proteins in a cell and run numerous tests to see what each one does. In a recent paper in Cell, however, researchers at the Dana-Farber Cancer Institute in Boston and their collaborators reveal the results of just such an effort. They found that in many cases, proteins made by a single gene are no more alike in their behavior than proteins made by completely different genes. Sibling proteins often act like strangers. It’s an insight that opens up an interesting new set of possibilities for thinking about how the cell — and the human body — functions.
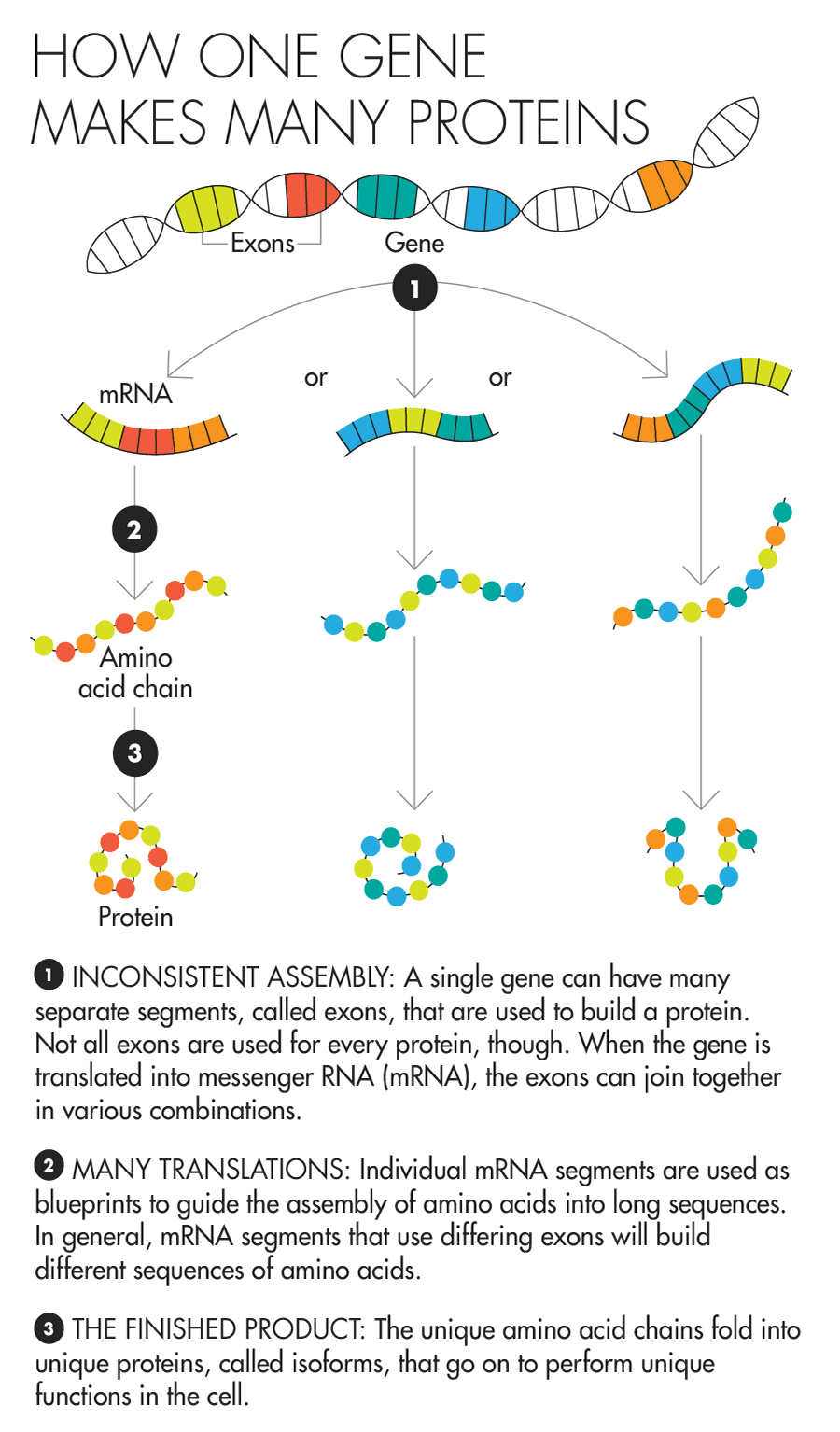
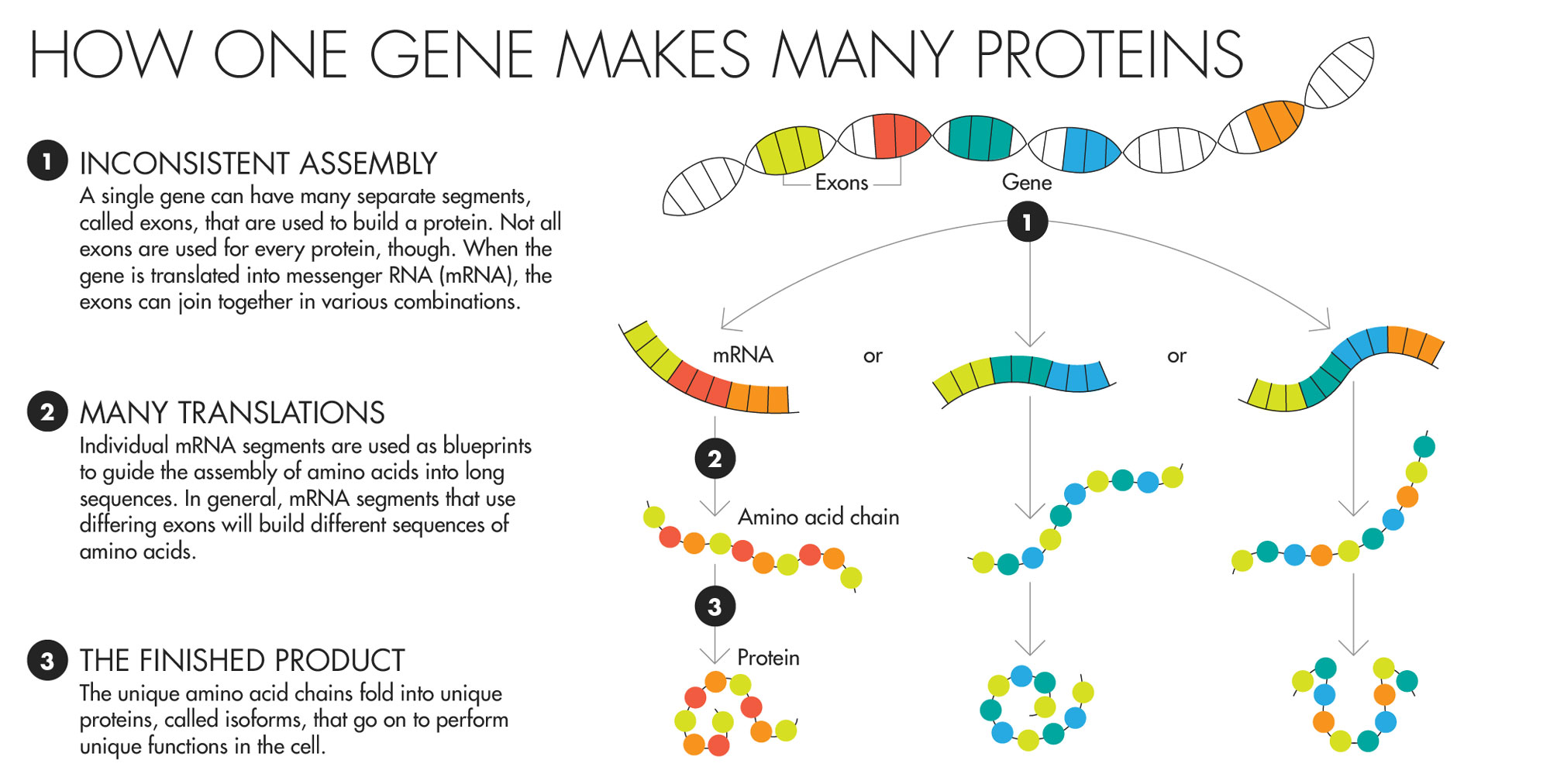
Lucy Reading-Ikkanda for Quanta Magazine
Proteins transact much of a cell’s daily business. Messages are sent from one part of the cell to another, for instance, by a protein bucket brigade — one attaches to another, which then switches on another, which then modifies another, and so on, culminating in a string of alterations that delivers the message. A protein’s particular shape helps determine what it can attach to and therefore what it can do. Finding out which proteins another protein will stick to is often the first step in understanding its role in the cell.
Marc Vidal, a biologist at Dana-Farber, has a long history of tracing such protein partnerships on a grand scale. His lab looks to see how large numbers of proteins interact with one another and how those interactions might change in someone with a disease. But it can be frustrating to do this when you aren’t sure whether you should assume that proteins from the same gene do the same thing. Even if we perfectly understand a particular genome sequence, “we still don’t have a perfect knowledge of the components that are encoded by the genome,” Vidal said. “And the reason is that the good old rules don’t hold.”
To see just how often the old rules might be broken, the Vidal lab and their collaborators gathered a set of proteins made from about 1,500 genes — about 8 percent of our total complement. They sorted out which proteins came from the same genes, finding that about 500 of the genes made at least two. Then they ran multiple tests in which each of the proteins was given the chance to attach to more than 15,000 other proteins often found in the cell. Finally, they compared each protein’s results to those of its sibling proteins — all those proteins made by the same gene. How often did sibling proteins attach to the same partners? How often did they not?
The answer was rather unexpected. “It was so striking,” said David Hill, a scientist at Dana-Farber, that he thought, “This can’t be right; we’ve got to figure out what we did wrong.” But the results held up to prodding. They found that 61 percent of sibling protein pairs share some but not all of their interactions. Moreover, nearly one in five of all sibling protein pairs had nothing in common. Comparing the proteins in their data set with proteins made by separate genes, the team found that in many cases the sibling proteins’ interactions were as different as if they’d had totally unrelated origins.
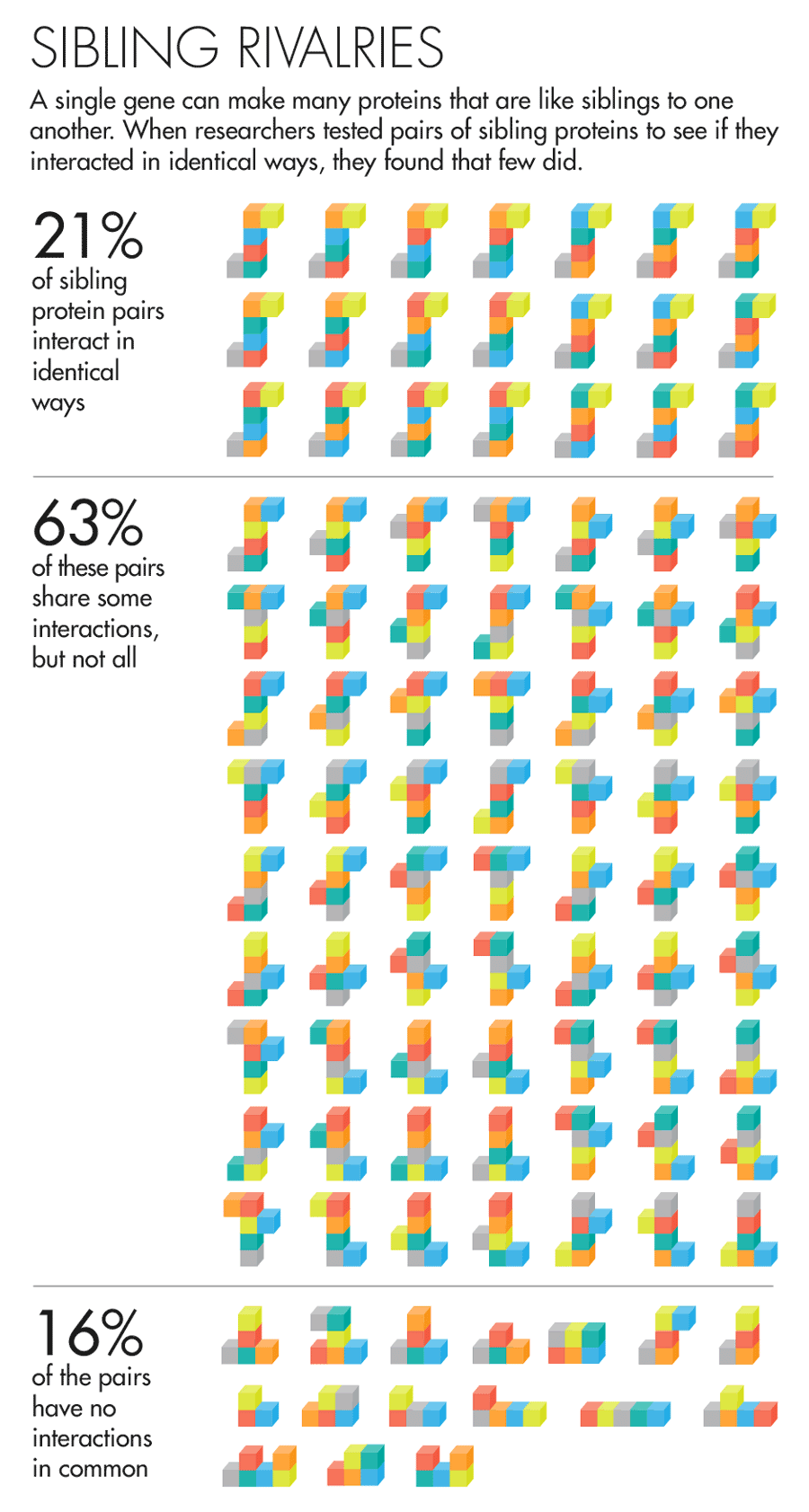
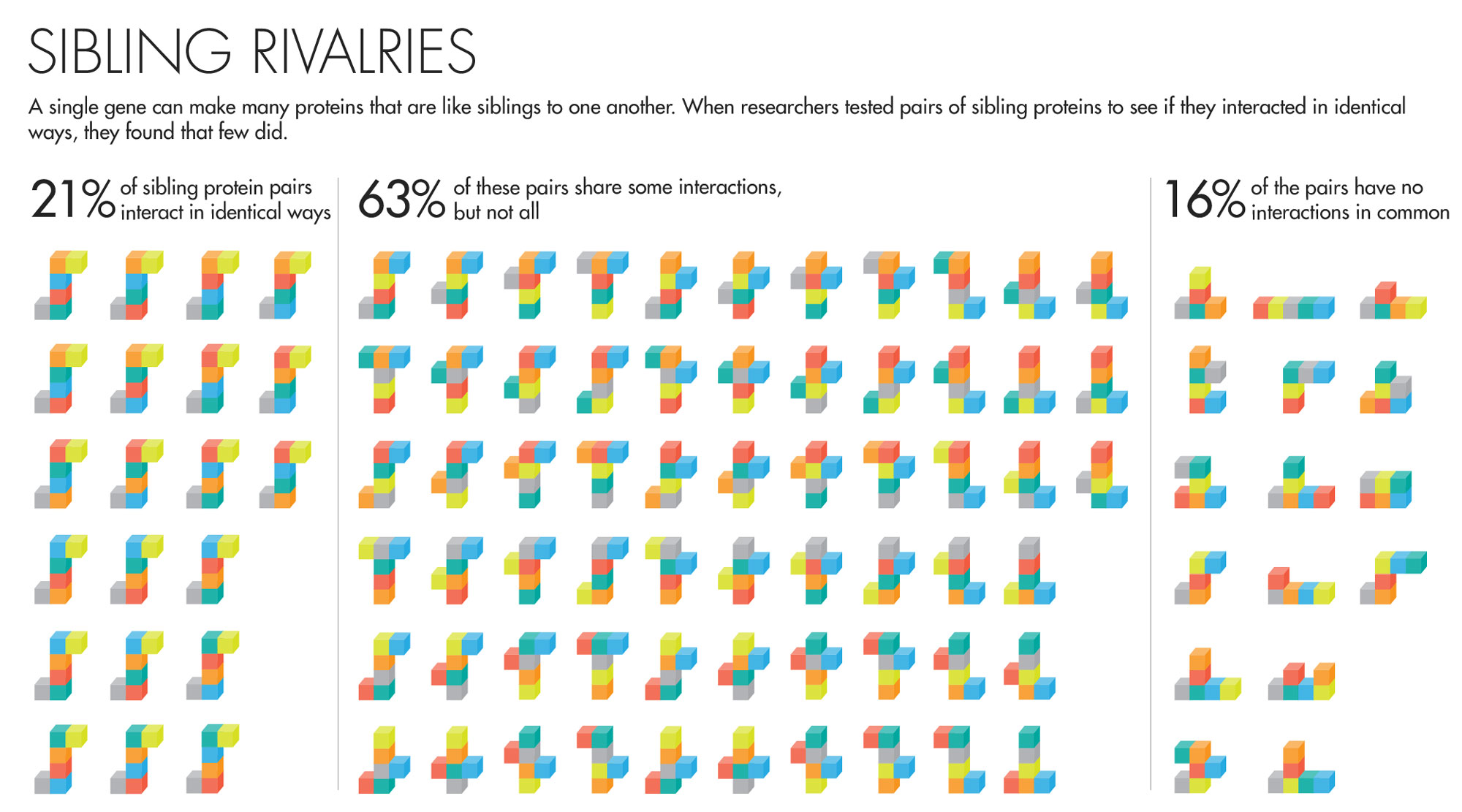
Lucy Reading-Ikkanda for Quanta Magazine
Because this paper suggests that different functions for proteins from the same gene are relatively common, it implies that the phenomenon probably matters for the everyday life of the cell, said Neil Kelleher, a biologist at Northwestern University who was not involved with the research. “We don’t know how much of the complexity of cells and tissues in our body arises from this,” he said. But it’s possible these different proteins could be part of what’s behind the distinct cell types in the body. Perhaps lung cells prefer to make one protein, while another protein predominates in a heart cell.
Some diseases might have their roots in one protein dominating where it shouldn’t. For example, a 2014 paper implies that certain alternative forms of proteins may play a role in autism. Additionally, the new research suggests that when researchers are trying to understand the biological underpinnings of a disease, they should not assume that it will be enough to pinpoint the genes involved. If a gene makes multiple proteins, biologists will need to deduce which protein is responsible for the problem.
Yet it remains to be seen just how relevant the new findings will be for understanding typical cell behavior. Stefan Stamm, a biologist at the University of Kentucky, notes that the study does not assess whether every observed protein interaction happens on a regular basis in real life. Previous work suggests that some of these proteins exist only in small numbers in the wild. But Stamm agrees that we’re ignorant of a lot of the variety in the world of proteins. “Personally, I think that there are more [alternative versions] than are being reported,” he said.
Hill estimates that the team has upped the number of genes known to make multiple proteins substantially. But “this is still the tip of the iceberg,” he notes. The team started with just 1,500 genes. Looking at 10,000 — half of all human genes — would make it clearer how widespread multiprotein genes are. The team might also choose to look deeper at a small handful of genes, getting a better picture of what their multiplicity of proteins is doing and observe how important they actually are within the cell. Either way, there is still much more to know.
The complexity implied by this finding may feel slightly overwhelming: How can we begin to unpack the biology of cells and tissues if there are so many different proteins coming from genes that, not all that long ago, people thought could make only a single one? But Kelleher said that in a sense, these results are reassuring. Theoretically, taking into account all the ways that a recipe provided by a gene could be interpreted — all the chances to substitute salt for sugar, say, or all the times when baking soda could replace baking powder — there could be up to 50 different proteins per gene.
This study suggests that in reality, only a small fraction of those possibilities are made. And only some of these proteins behave differently from one another. “People go ‘Oh my God, it’s so vast.’ But we can measure this,” he said hopefully. “It’s not so complicated as to be unknowable.”
This article was reprinted on TheAtlantic.com.