How Neanderthal DNA Helps Humanity

Native Tibetans make use of a gene derived from Denisovans to stay healthy at high altitudes.
Introduction
Early human history was a promiscuous affair. As modern humans began to spread out of Africa roughly 50,000 years ago, they encountered other species that looked remarkably like them — the Neanderthals and Denisovans, two groups of archaic humans that shared an ancestor with us roughly 600,000 years earlier. This motley mix of humans coexisted in Europe for at least 2,500 years, and we now know that they interbred, leaving a lasting legacy in our DNA. The DNA of non-Africans is made up of roughly 1 to 2 percent Neanderthal DNA, and some Asian and Oceanic island populations have as much as 6 percent Denisovan DNA.
Over the last few years, scientists have dug deeper into the Neanderthal and Denisovan sections of our genomes and come to a surprising conclusion. Certain Neanderthal and Denisovan genes seem to have swept through the modern human population — one variant, for example, is present in 70 percent of Europeans — suggesting that these genes brought great advantage to their bearers and spread rapidly.

The Downside to Neanderthal DNA
“In some spots of our genome, we are more Neanderthal than human,” said Joshua Akey, a geneticist at the University of Washington. “It seems pretty clear that at least some of the sequences we inherited from archaic hominins were adaptive, that they helped us survive and reproduce.”
But what, exactly, do these fragments of Neanderthal and Denisovan DNA do? What survival advantage did they confer on our ancestors? Scientists are starting to pick up hints. Some of these genes are tied to our immune system, to our skin and hair, and perhaps to our metabolism and tolerance for cold weather, all of which might have helped emigrating humans survive in new lands.
“What allowed us to survive came from other species,” said Rasmus Nielsen, an evolutionary biologist at the University of California, Berkeley. “It’s not just noise, it’s a very important substantial part of who we are.”
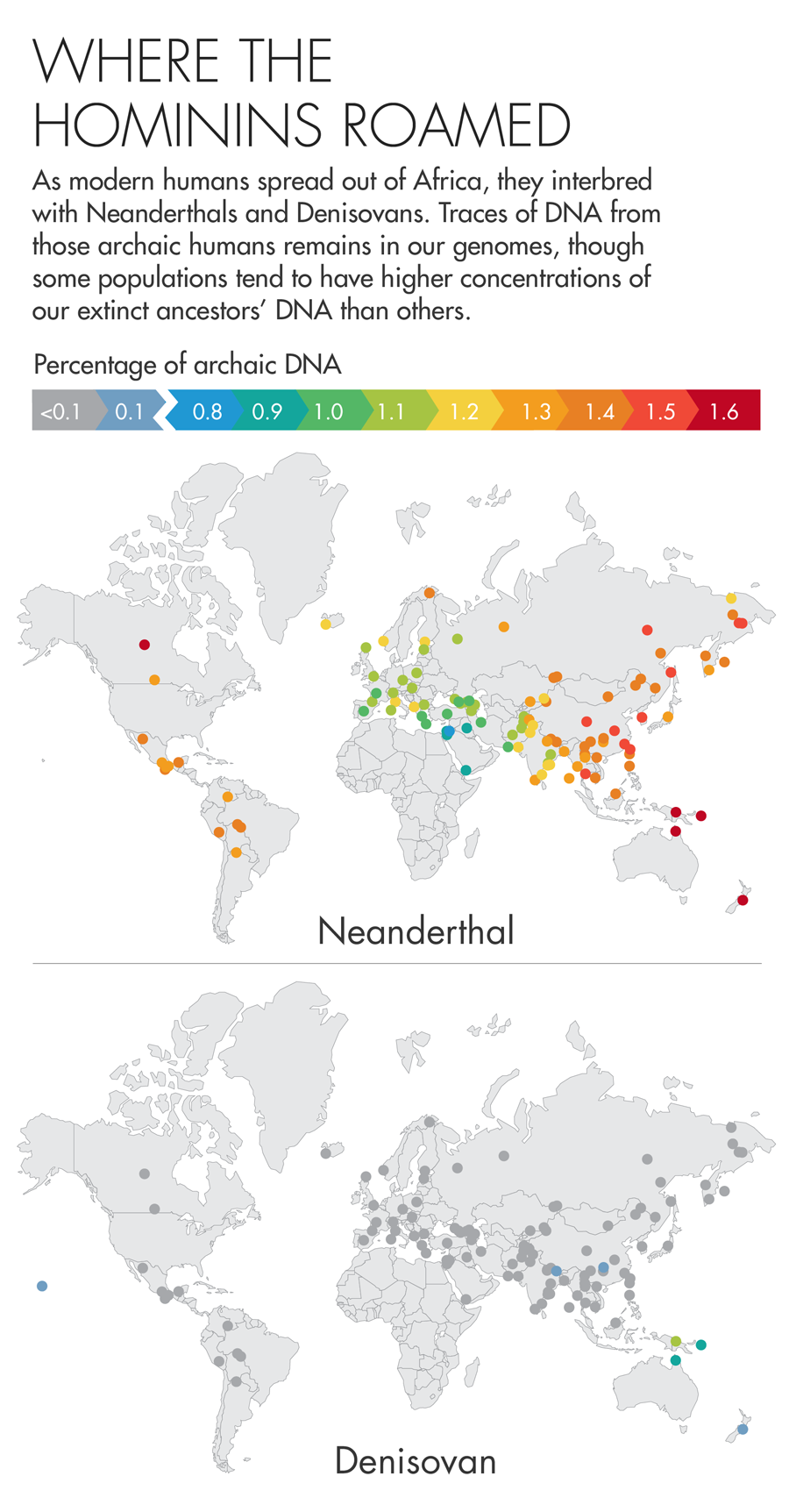
Illustration by Lucy Reading-Ikkanda for Quanta Magazine, based on a map by Sriram Sankararaman.
The Neanderthal Within
The Tibetan plateau is a vast stretch of high-altitude real estate isolated by massive mountain ranges. The scant oxygen at 14,000 feet — roughly 40 percent lower than the concentrations at sea level — makes it a harsh environment. People who move there suffer higher rates of miscarriage, blood clots and stroke on account of the extra red blood cells their bodies produce to feed oxygen-starved tissue. Native Tibetans, however, manage just fine. Despite the meager air, they don’t make as many red blood cells as the rest of us would at those altitudes, which helps to protect their health.
In 2010, scientists discovered that Tibetans owe their tolerance of low oxygen levels in part to an unusual variant in a gene known as EPAS1. About 90 percent of the Tibetan population and a smattering of Han Chinese (who share a recent ancestor with Tibetans) carry the high-altitude variant. But it’s completely absent from a database of 1,000 human genomes from other populations.
In 2014, Nielsen and colleagues found that Tibetans or their ancestors likely acquired the unusual DNA sequence from Denisovans, a group of early humans first described in 2010 that are more closely related to Neanderthals than to us. The unique gene then flourished in those who lived at high altitudes and faded away in descendants who colonized less harsh environments. “That’s one of the most clear-cut examples of how [interbreeding] can lead to adaptation,” said Sriram Sankararaman, a geneticist and computer scientist at the University of California, Los Angeles.
The idea that closely related species can benefit from interbreeding, known in evolutionary terms as adaptive introgression, is not a new one. As a species expands into a new territory, it grapples with a whole new set of challenges — different climate, food, predators and pathogens. Species can adapt through traditional natural selection, in which spontaneous mutations that happen to be helpful gradually spread through the population. But such mutations strike rarely, making it a very slow process. A more expedient option is to mate with species that have already adapted to the region and co-opt some of their helpful DNA. (Species are traditionally defined by their inability to mate with one another, but closely related species often interbreed.)
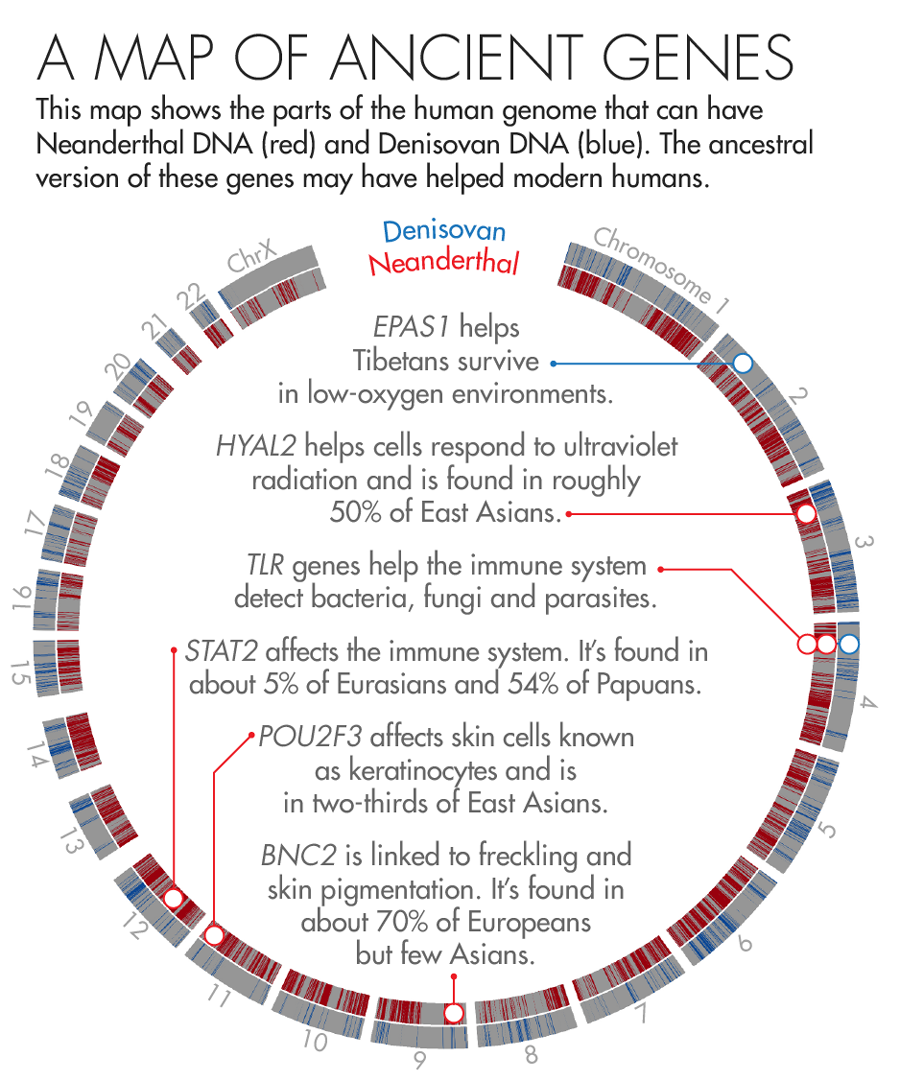
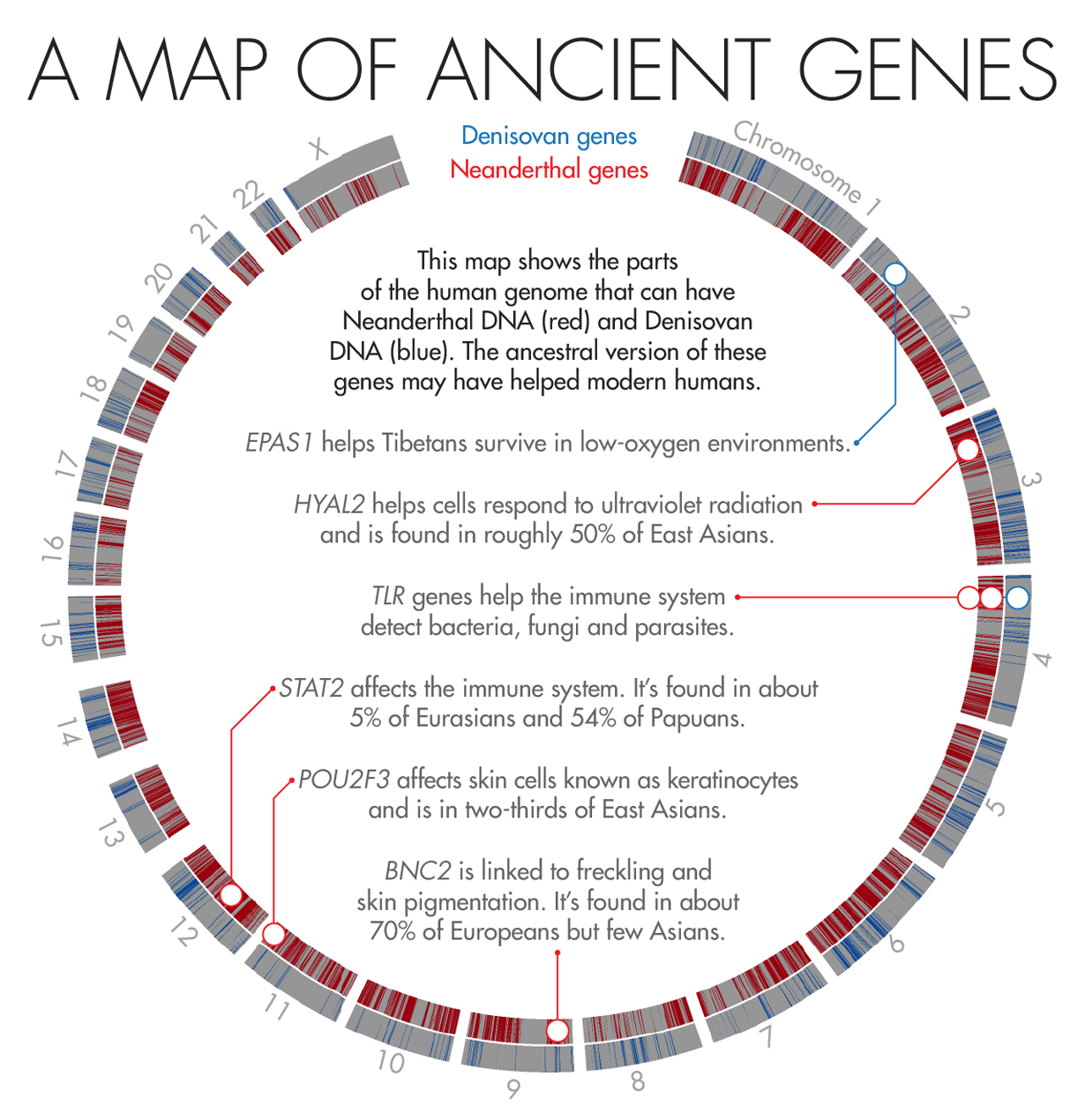
Illustration by Lucy Reading-Ikkanda for Quanta Magazine, based on a map by Sriram Sankararaman
This phenomenon has been well documented in a number of species, including mice that adopted other species’ tolerance to pesticides and butterflies that appropriated other species’ wing patterning. But it was difficult to study adaptive introgression in humans until the first Neanderthal genome was sequenced in 2010, providing scientists with hominin DNA to compare to our own.
Neanderthals and Denisovans would have been a good source of helpful DNA for our ancestors. They had lived in Europe and Asia for hundreds of thousands of years — enough time to adjust to the cold climate, weak sun and local microbes. “What better way to quickly adapt than to pick up a gene variant from a population that had probably already been there for 300,000 years?” Akey said. Indeed, the Neanderthal and Denisovan genes with the greatest signs of selection in the modern human genome “largely have to do with how humans interact with the environment,” he said.
To find these adaptive segments, scientists search the genomes of contemporary humans for regions of archaic DNA that are either more common or longer than expected. Over time, useless pieces of Neanderthal DNA — those that don’t help the carrier — are likely to be lost. And long sections of archaic DNA are likely to be split into smaller segments unless there is selective pressure to keep them intact.
In 2014, two groups, one led by Akey and the other by David Reich, a geneticist at Harvard Medical School, independently published genetic maps that charted where in our genomes Neanderthal DNA is most likely to be found. To Akey’s surprise, both maps found that the most common adaptive Neanderthal-derived genes are those linked to skin and hair growth. One of the most striking examples is a gene called BNC2, which is linked to skin pigmentation and freckling in Europeans. Nearly 70 percent of Europeans carry the Neanderthal version.
Scientists surmise that BNC2 and other skin genes helped modern humans adapt to northern climates, but it’s not clear exactly how. Skin can have many functions, any one of which might have been helpful. “Maybe skin pigmentation, or wound healing, or pathogen defense, or how much water loss you have in an environment, making you more or less susceptible to dehydration,” Akey said. “So many potential things could be driving this — we don’t know what differences were most important.”
Surveillance System
One of the deadliest foes that modern humans had to fight as they ventured into new territories was also the smallest — novel infectious diseases for which they had no immunity. “Pathogens are one of the strongest selective forces out there,” said Janet Kelso, a bioinformatician at the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany.
Earlier this year, Kelso and collaborators identified a large stretch of Neanderthal DNA — 143,000 DNA base-pairs long — that may have played a key role in helping modern humans fight off disease. The region spans three different genes that are part of the innate immune system, a molecular surveillance system that forms the first line of defense against pathogens. These genes produce proteins called toll-like receptors, which help immune cells detect foreign invaders and trigger the immune system to attack.
Modern humans can have several different versions of this stretch of DNA. But at least three of the variants appear to have come from archaic humans — two from Neanderthals and one from Denisovans. To figure out what those variants do, Kelso’s team scoured public databases housing reams of genomic and health data. They found that people carrying one of the Neanderthal variants are less likely to be infected with H. pylori, a microbe that causes ulcers, but more likely to suffer from common allergies such as hay fever.
Kelso speculates that this variant might have boosted early humans’ resistance to different kinds of bacteria. That would have helped modern humans as they colonized new territories. Yet this added resistance came at a price. “The trade-off for that was a more sensitive immune system that was more sensitive to nonpathogenic allergens,” said Kelso. But she was careful to point out that this is just a theory. “At this point, we can hypothesize a lot, but we don’t know exactly how this is working.”
Most of the Neanderthal and Denisovan genes found in the modern genome are more mysterious. Scientists have only a vague idea of what these genes do, let alone how the Neanderthal or Denisovan version might have helped our ancestors. “It’s important to understand the biology of these genes better, to understand what selective pressures were driving the changes we see in present-day populations,” Akey said.
A number of studies like Kelso’s are now under way, trying to link Neanderthal and Denisovan variants frequently found in contemporary humans with specific traits, such as body-fat distribution, metabolism or other factors. One study of roughly 28,000 people of European descent, published in Science in February, matched archaic gene variants with data from electronic health records. Overall, Neanderthal variants are linked to higher risk of neurological and psychiatric disorders and lower risk of digestive problems. (That study didn’t focus on adaptive DNA, so it’s unclear how the segments of archaic DNA that show signs of selection affect us today.)
At present, much of the data available for such studies is weighted toward medical problems — most of these databases were designed to find genes linked to diseases such as diabetes or schizophrenia. But a few, such as the UK Biobank, are much broader, storing information on participants’ vision, cognitive test scores, mental health assessments, lung capacity and fitness. Direct-to-consumer genetics companies also have large, diverse data sets. For example, 23andMe analyzes users’ genetics for clues about ancestry, health risk and other sometimes bizarre traits, such as whether they have a sweet tooth or a unibrow.
Of course, not all the DNA we got from Neanderthals and Denisovans was good. The majority was probably detrimental. Indeed, we tend to have less Neanderthal DNA near genes, suggesting that it was weeded out by natural selection over time. Researchers are very interested in these parts of our genomes where archaic DNA is conspicuously absent. “There are some really big places in the genome with no Neanderthal or Denisovan ancestry as far as we can see — some process is purging the archaic material from these regions,” Sankararaman said. “Perhaps they are functionally important for modern humans.”
This article was reprinted on TheAtlantic.com.