A ‘Lobby’ Where a Molecule Mob Tells Genes What to Do
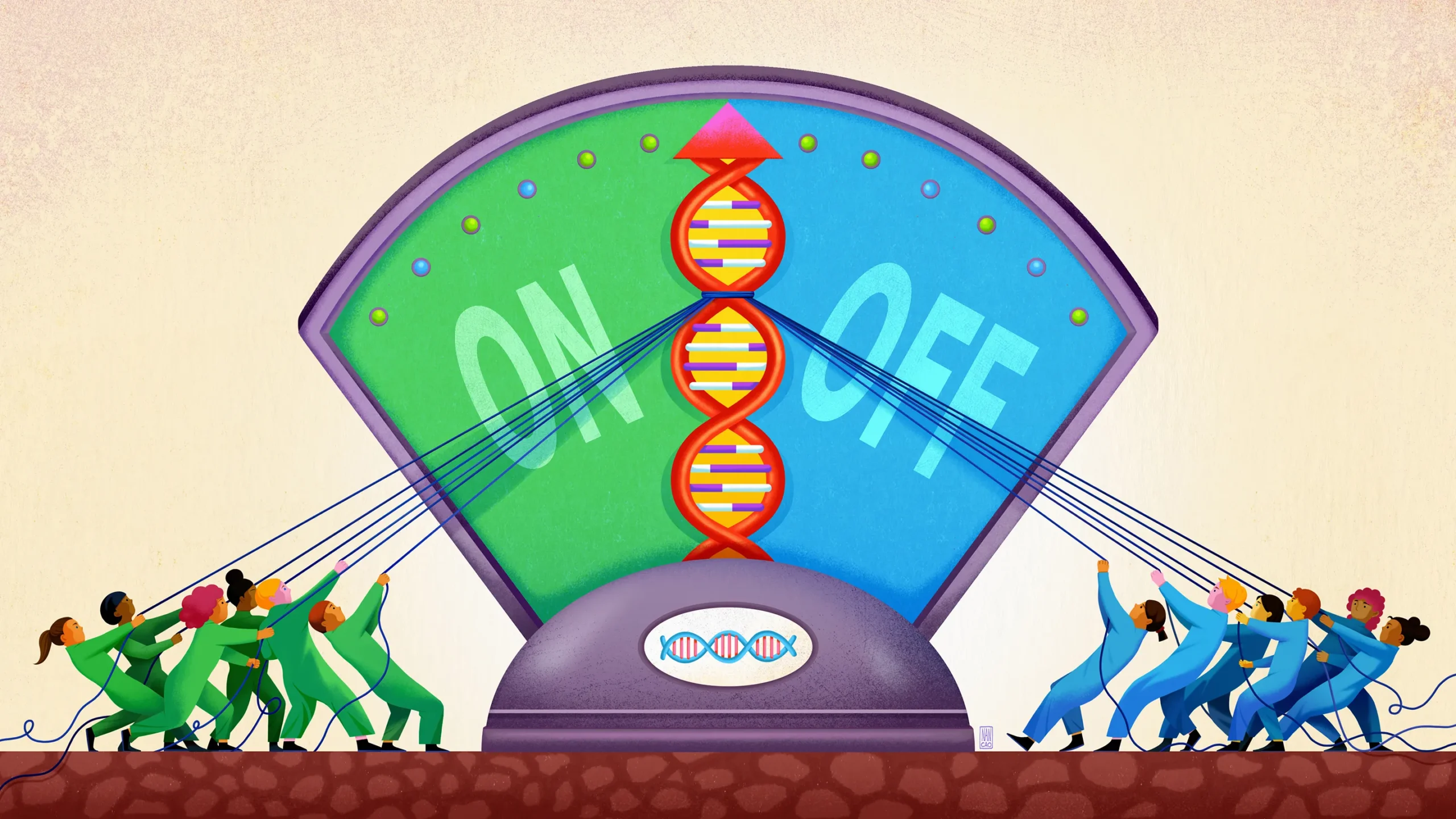
Gene regulation in humans and other complex organisms is an elaborate process involving many molecular factors that can have opposing effects.
Nan Cao for Quanta Magazine
Introduction
The discovery during the Human Genome Project in the early 2000s that we humans have only about 20,000 protein-coding genes — about as many as the tiny soil-dwelling nematode worm, and less than half as many as the rice plant — came as a shock. That blow to our pride was softened, though, by the idea that the human genome is rich in regulatory connections. Our genes interact in a dense network, in which pieces of DNA and the molecules they encode (RNA and proteins) control the “expression” of other genes, influencing whether they make their respective RNA and proteins. To understand the human genome, we needed to understand this process of gene regulation.
That task, however, is proving to be much harder than decoding the sequence of the genome.
Initially, it was suspected that gene regulation was a simple matter of one gene product acting as an on/off switch for another gene, in digital fashion. In the 1960s, the French biologists François Jacob and Jacques Monod first elucidated a gene regulatory process in mechanistic detail: In Escherichia coli bacteria, when a repressor protein binds to a certain segment of DNA, it blocks the transcription and translation of an adjacent suite of genes that encode enzymes for digesting the sugar lactose. This regulatory circuit, which Monod and Jacob dubbed the lac operon, has a neat, transparent logic.
But gene regulation in complex metazoans — animals like humans, with complex eukaryotic cells — doesn’t generally seem to work this way. Instead, it involves a gang of molecules, including proteins, RNAs and pieces of DNA from throughout a chromosome, that somehow collaborate to control the expression of a gene.
It’s not just that this regulatory process in eukaryotes has more players than is typically seen in bacteria and other simple, prokaryotic cells; it seems to be a categorically different process, and a hazier one.
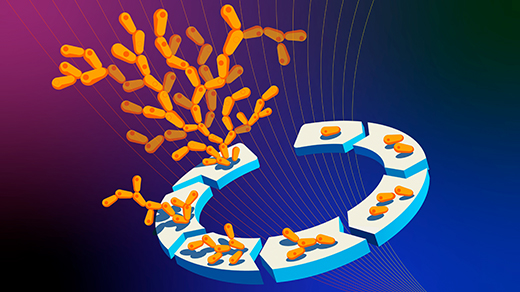
A team at Stanford University, led by the biophysicist and bioengineer Polly Fordyce, seems now to have uncovered a component of this fuzzy mode of gene regulation. Their work, published last September in Science, suggests that the DNA near a gene acts as a kind of shallow well for trapping diverse regulatory molecules, keeping them ready for action so that, when needed, they can add their voice to the decision about whether to activate the gene.

The biophysicist Polly Fordyce and the members of her laboratory at Stanford University may have discovered how short, highly repetitive sequences of DNA near genes play a role in the regulation of transcription.
Stanford Bioengineering Communications Team
These regulatory wells are made from decidedly odd stretches of DNA. They consist of sequences in which a short stretch of DNA, from one to six base pairs long, repeats many times over. Tens of copies of these “short tandem repeats” (STRs) can be strung together in these sequences, like the same little “word” written again and again.
STRs are abundant in the human genome: They comprise about 5% of all our DNA. They were once thought to be classic examples of “junk” DNA because a repetitive DNA “text” made up only of STRs can’t hold nearly as much meaningful information as, say, the irregular sequence of letters that make up a sentence in this article.
And yet, STRs are clearly not insignificant: They have been linked to ailments such as Huntington’s disease, spinobulbar muscular atrophy, Crohn’s disease and some cancers. Over the past couple of decades, evidence has accumulated that they can somehow enhance or inhibit gene regulation. The mystery was how they could be so powerful with so little information content.
Complex Controls for Complex Cells
To understand how STRs fit into the big picture of gene regulation, let’s take a step back. Genes are routinely flanked by pieces of DNA that don’t encode RNA or protein but have regulatory functions. Bacterial genes have “promoter” regions where polymerase enzymes can bind to begin the transcription of adjacent DNA into RNA. They also routinely have “operator” regions, where repressor proteins can bind to block transcription, turning a gene off, as in the lac operon.
In humans and other eukaryotes, the regulatory sequences can be more numerous, various — and perplexing. Regions called enhancers, for example, affect the probability that a gene will be transcribed. Enhancers are often the targets for proteins called transcription factors, which can bind to boost or inhibit gene expression. Weirdly, some enhancers are tens of thousands of base pairs away from the genes they regulate, and are only brought close to them through the physical rearrangement of the loops of DNA in a packed chromosome.
Merrill Sherman/Quanta Magazine
Eukaryotic gene regulation typically involves these many diverse regulatory blocks of DNA, along with one or more transcription factors and other molecules, all gathering around a gene like a committee convened to decide what it should do. They congregate in a loose, dense cluster.
Often, the molecular participants also don’t seem to interact through the highly selective “lock and key” pairings common in molecular biology. They are instead much less choosy, interacting rather weakly and unselectively, as though wandering around and striking up brief conversations with one another.
In fact, how transcription factors bind to DNA in eukaryotes has been something of a mystery. It was long assumed that some part of a transcription factor must closely match a binding “motif” sequence in the DNA, like the pieces of a jigsaw puzzle. But although some such motifs have been identified, their presence doesn’t always correlate very well with where scientists find transcription factors sticking to DNA in cells. Sometimes transcription factors linger in regions without any motifs, while some motifs that seem as if they should strongly bind transcription factors remain empty.
“Traditionally in genomics, the goal has been to classify genomic sites in a [binary] way as either ‘bound’ or ‘unbound’” by transcription factors, Fordyce said. “But the picture is way more nuanced than that.” The individual members of those gene regulatory “committees” don’t seem to be invariably present for or absent from their meetings, but rather have different probabilities of being there or not.
The tendency of gene regulation in eukaryotes to rely on so many diverse weak interactions among large molecular complexes “is one of the things that makes it notoriously difficult to get a handle on theoretically,” said the biophysicist Thomas Kuhlman of the University of California, Riverside, who wrote a commentary on the Fordyce lab’s paper for Science. It’s a profound puzzle how, out of this seemingly chaotic process, precise decisions about turning genes on and off emerge.
Beyond the mysterious fuzzy logic of that decision process, there’s also the question of how all the committee members even find their way to the right room — and then stay there. Molecules generally move around the cell by diffusion, buffeted by all the other surrounding molecules, such as water, and wandering in random directions. We might expect these loose committees to drift apart too quickly to do their regulatory job.
That, Fordyce and her colleagues think, is where the STRs come in. STRs are strikingly common within enhancer sites on DNA. In their paper, the researchers argue that the STRs act as sticky patches that convene transcription factors and stop them from straying.
Fine-Tuning the Stickiness
Fordyce’s group systematically investigated how differences in STR sequence influence the sticking of transcription factors to a binding motif. They looked at two factors — one from yeast, one from humans — that stick to a particular six-base motif. The researchers measured both the strength (or affinity) of that binding and the rate at which the transcription factors become stuck and unstuck (kinetics) when the motif is flanked by an STR instead of a random sequence. For comparison, they looked at how readily the factors bind to the STR alone and to a wholly random DNA sequence.
“One of the biggest challenges of this field is to disentangle the myriad variables that impact [transcription factor] binding at a specific position of the genome,” said David Suter, a molecular biologist at the Swiss Federal Institute of Technology Lausanne in Switzerland. DNA shape, proximity to other DNA segments and physical tension in the DNA molecules can all play a role in transcription factor binding. The values of these parameters probably differ at every position in the genome, and maybe also between cell types and within a single cell over time at a given position. “This is a vast space of unknown variables that are very hard to quantify,” Suter said.

“The repetitive nature of an STR amplifies the weak effect of any single binding site that it is made of,” said Connor Horton, a coauthor on the paper from Fordyce’s lab.
Justin Shumate
That’s why well-controlled experiments like those of the Stanford team are so useful, Kuhlman added. Usually, when researchers need to measure weak interactions like these, they have two choices: They can make a few very detailed, extremely precise measurements and generalize from them, or they can take a great many quick-and-dirty measurements and use mathematically complex statistical methods to deduce results. But Fordyce and her colleagues, Kuhlman said, used an automated, microfluidic chip-based procedure to take precise measurements during high-throughput experiments “to get the best of both worlds.”
The Stanford team found that different STR sequences can alter the binding affinities of transcription factors to DNA by as much as a factor of 70; they sometimes have more impact on transcription factor binding than changing the sequence of the binding motif itself. And the effects were different for the two different transcription factors they looked at.
So STRs seem able to fine-tune the ability of transcription factors to dock at a DNA site and thus to regulate a gene. But how, exactly?
A Waiting Room Near a Gene
The researchers figured that the part of a transcription factor that binds DNA might interact weakly with an STR, with the exact strength of that affinity depending on the STR sequence. Because such binding is feeble, it won’t have much specificity. But if a transcription factor is loosely grasped and released by an STR again and again, the cumulative effect is to keep the transcription factor in the vicinity of the gene so that it is more likely to bind securely to the motif region if needed.
Fordyce and her colleagues predicted that STRs thus act as a “lobby” or well where transcription factors can gather, however transiently, near a regulatory binding site. “The repetitive nature of an STR amplifies the weak effect of any single binding site that it is made of,” said Connor Horton, the first author on the study, who is now a doctoral student at the University of California, Berkeley.
Merrill Sherman/Quanta Magazine
Conversely, he added, some STRs can also act to pull transcription factors away from regulatory sequences, soaking up transcription factors elsewhere like a sponge. In this way, they can inhibit gene expression.
The work, Suter said, “shows convincingly that STRs directly impact binding of transcription factors in vitro.” What’s more, the Stanford team used a machine learning algorithm to show that the effects seen in their in vitro experiments also seem to be occurring in living cells (that is, in vivo).
But Robert Tjian, a biochemist at Berkeley and an investigator at the Howard Hughes Medical Institute, thinks it may be too early to be sure what influence a given STR-transcription factor combination has on gene expression in real cells.
Tjian, Xavier Darzacq and their colleagues in the lab they run together at Berkeley agree that STRs seem to offer a way of concentrating transcription factors near gene regulatory sites. Yet without knowing how close the factors need to be to activate transcription, it’s difficult to understand the functional significance of that result. Tjian said he would like to see whether introducing an STR into a living cell predictably influences the expression of a target gene. At present, he said, he is “not persuaded that STRs are necessarily going to be a major aspect of [regulatory] mechanisms in vivo.”
A Combinatorial Grammar
One lingering puzzle is how such a mechanism reliably provides the type of precise gene regulation that cells need, since both the strength and the selectivity of transcription factor binding within the STR wells are weak. Fordyce thinks that such specificity of influence could come from many sources — not just from differences in the STR sequences but also from cooperative interactions between transcription factors and other proteins involved in regulation.
Given all that, Horton said, it’s not clear that it will be straightforward to predict the effect of a given STR-transcription factor combination on the expression of a gene. The logic of the process is fuzzy indeed. And the “grammar” of the influence is probably combinatorial, Horton added: The outcome depends on different combinations of transcription factors and other molecules.
The Stanford team thinks that perhaps 90% of transcription factors are sensitive to STRs, but that there are many more types of transcription factors in the human genome than there are types of STRs. “Mutating an STR sequence might affect the binding of 20 different transcription factors in that cell type, leading to an overall decrease in transcription of that nearby gene without implicating any specific transcription factor,” Horton said.
So in effect, the Stanford team agrees with Tjian that gene regulation in living cells isn’t going to be driven by a single, simple mechanism. Rather, transcription factors, their DNA binding sites, and other regulatory molecules may assemble into dense gatherings that exert their influence collectively.
“There are now multiple examples that support the idea that DNA elements can crowd transcription factors to the point where they form condensates with cofactors,” said Richard Young, a cell biologist at the Whitehead Institute of the Massachusetts Institute of Technology. Enhancers bind many transcription factors to produce that crowding. STRs may be an ingredient that helps muster transcription factors to cluster near a gene, but they won’t be the whole story.
Why regulate genes in this complicated manner, rather than relying on the kind of strong and specific interactions between regulatory proteins and DNA sites that dominate in prokaryotes? It’s possible that such fuzziness is what made large complex metazoans possible at all.
To be viable species, organisms need to be able to evolve and adapt to changing circumstances. If our cells relied on some huge yet tightly prescribed network of gene regulatory interactions, it would be difficult to make any changes to it without disrupting the whole contraption, just as a Swiss watch will seize up if we remove (or even slightly displace) any of its myriad cogwheels. If the regulatory molecular interactions are loose and rather unspecific, however, there is useful slack in the system — just as a committee can generally come to a good decision even if one of its members is out sick.
Fordyce notes that in prokaryotes like bacteria, it may be relatively easy for transcription factors to find their binding sites because the genome to be searched is smaller. But that gets harder as the genome gets bigger. In the big genomes of eukaryotes, “you can no longer tolerate the risk that you will become transiently stuck at a ‘wrong’ binding site,” Fordyce said, because that would compromise the ability to respond quickly to changing environmental conditions.
Moreover, STRs themselves are highly evolvable. A lengthening or shortening of their sequence, or an alteration to the size and depth of the “transcription factor well,” can occur easily through mishaps in DNA replication or repair, or through sexual recombination of the chromosomes. To Fordyce, it suggests that STRs “may therefore serve as the raw material for evolving new regulatory elements and fine-tuning existing regulatory modules for sensitive transcriptional programs,” such as those governing the development of animals and plants.
The Power of Weak Interactions
Such considerations are leading molecular biologists to pay much more attention to weak and relatively unselective interactions in the genome. Many of these involve proteins that, instead of having a fixed and precise structure, are loose and floppy — “intrinsically disordered,” as biochemists put it. If proteins only worked through rigid structural domains, Young explained, it would constrain not only how well regulatory systems could evolve but also the kinds of dynamic regulation seen in life. “You won’t find a living organism — or even a virus — functioning with only stable structural elements like those in a Swiss watch,” Young said.
Perhaps evolution just stumbled on STRs as a component of such a complex but ultimately more effective solution to gene regulation in eukaryotes. STRs themselves may arise in several ways — for example, through errors in DNA replication or the activity of DNA segments called transposable elements that make copies of themselves throughout the genome.
“It just so happened that the resulting emergent weak interactions between proteins and the repetitive sequences was something that could … provide selective advantage to the cells where it occurred,” Kuhlman said. His guess is that this fuzziness was probably forced upon eukaryotes, but that “they were subsequently able to exploit [it] for their own benefit.” Bacteria and other prokaryotes can rely on well-defined “digital” regulatory logic because their cells tend to exist in only a few simple, distinct states, such as moving around and replicating.
But the different cell states for metazoans are “much more complex and sometimes close to a continuum,” Suter said, so they are better served by fuzzier “analog” regulation.
“The gene regulatory systems in bacteria and eukaryotes do seem to have diverged quite substantially,” Tjian agreed. While Monod is said to have once remarked that “what is true for E. coli is true for the elephant,” it seems that isn’t always so.