Her Machine Learning Tools Pull Insights From Cell Images

Today, biomedical researchers can efficiently classify thousands of cells in microscopy images by using machine learning for image-based profiling. The computational biologist Anne Carpenter is a pioneer in the development of these automated tools.
Bearwalk Cinema
Introduction
You can’t judge a book by its cover, or so we’re taught about people. For cells, however, that’s surprisingly less true. Using machine learning methods similar to those that enable computers to recognize faces, biologists can characterize individual cells in stacks of microscopy images. By measuring thousands of visualizable cellular properties — the distribution of a tagged protein, the shape of the nucleus, the number of mitochondria — computers can mine images of cells for patterns that identify their cell type and disease-associated traits. This kind of image-based profiling is speeding up drug discovery by improving screening for compounds that desirably modify cells’ characteristics.
Anne Carpenter, a computational biologist and senior director of the Imaging Platform of the Broad Institute of the Massachusetts Institute of Technology and Harvard University, is a pioneer of this approach to research. She developed CellProfiler, a widely used open-source software for measuring phenotypes (sets of observable traits) from cell images. It has been cited in more than 12,000 publications since its release in 2005.
It started out as a side project during her training as a cell biologist — what Carpenter calls “a little scrap of code to do a thing” that she needed, which over time grew into a toolbox that other researchers found useful, too. “By the time I got toward the end of my postdoc, I found that I would much rather help other people accomplish their cool biology by making the tools than pursue my own particular biological questions,” she said. “That’s why I ended up staying in computer science.”
A Massachusetts Academy of Sciences fellow, Carpenter has received a National Institutes of Health MIRA award, as well as a CAREER award from the National Science Foundation and a 2020 Women in Cell Biology Mid-Career Award from the American Society for Cell Biology, among other honors.
Carpenter spoke with Quanta Magazine about the joy of translating messy biology into computationally solvable problems, an ambitious effort to screen drugs for 200 diseases in a single well, and how researchers who are humble, curious and able to communicate with people outside their discipline can create a culture that improves the diversity of computational biology and machine learning. The interview has been condensed and edited for clarity.

Carpenter and the co-leader of her laboratory, Shantanu Singh, assembled a research team by focusing on the skills, curiosity and communication abilities of candidates. “Without explicitly trying, my lab has been much more diverse than average for a computational lab at a top-tier institution,” she said.
Bearwalk Cinema
Computer scientists have applied their skills in biology, but you took the less common path from biology into software engineering. What motivated you?
The transition was born out of necessity. During my cell biology doctorate work at the University of Illinois, Urbana-Champaign in the early 2000s, I was studying how chromatin, the complex of DNA and proteins in eukaryotic cells, responds to signals passed through the estrogen receptor. This required capturing thousands of microscopy images. It would have taken months to do manually. I decided that it would be great if I could figure out how to automate the microscope.
I had no formal training in computer science. It took about a month to figure out how to program the microscope, but that saved me two months of time manually collecting images in a really boring fashion.
It also created a new challenge: I now had a huge pile of images to analyze. I spent more months and months copying and pasting code, figuring that out as I went.
Once I got into playing with image analysis, though, I was hooked. It was so satisfying to be able to turn messy, qualitative biology into precise, quantitative numbers. I decided to seek a postdoc position where I could accelerate biology by working on high-throughput imaging.
In a recent essay you describe biology as “messy” but also “a logic puzzle.” Can you talk a bit more about that?
Biology is quite messy. It’s really hard to figure anything out. You would hope that A activates B, which activates C, and then C represses D, and so on. But in reality, there are so many weird, imprecise relationships — like feedbacks, multiple inputs, alternate pathways — going on in cells.
Yet I also believe biology is a logic puzzle. The best we can do is try to constrain the model system we’re testing. Then we can perturb it, measure inputs and outputs, and so on. We can turn biology into a less messy thing by imposing a lot of constraints on it.
During your postdoc at the Whitehead Institute, you started working on what eventually became CellProfiler. How did you go about that?
I had realized I needed some serious new code for my project, so I just dove in and learned some programming by trial and error. But I still needed help implementing some of the classical image-processing algorithms. I’d read a paper and say, “This is exactly what I need” — but I had no clue how to transform the paper’s equations into code.
I sent an email to the graduate student list at MIT’s Computer Science and Artificial Intelligence Laboratory and asked: “Does anybody want to help me? I have some fellowship money.” Thouis (Ray) Jones responded and, in one weekend, implemented the core algorithms. They were quite revolutionary and formed the core of why CellProfiler became so successful: It made those algorithms available to end users.
By quantifying phenotypic differences in a variety of cells on a large scale, CellProfiler can be used for “image-based profiling.” How did you hit on the idea for this?
People would come to us and say: “Here’s my fancy cell type. Here’s my special antibody to label some protein in the cell. Can you tell me how much of my protein is present in the nucleus?” Of course, with image analysis, we could measure whatever they asked for.
But looking at the images, I would say: “Did you also notice that the protein’s texture is changing? Or that it’s actually more at the edge of the nucleus than in the interior? And we see co-localization between this stain and that stain. And the overall shape of the cell is changing. Is that biologically meaningful?” There was so much information the biologists were leaving on the table!
That’s when I was inspired by a 2004 Science paper, where researchers carried out image-based profiling on cells treated with various sets of compounds. They showed that cells treated with functionally similar compounds tended to look alike — the compounds had a similar impact on the cell. It was electrifying. Could it really be that humble, beautiful images of cells carry enough quantitative information to tell us what drug the cells had been treated with? That paper really launched the field of image-based profiling.
What does this profiling involve?
We measure everything we can about the cell’s appearance. We’re building on the basic observation that a cell’s structure and overall appearance reflects its history — how it’s been treated by its environment. If images reflect the state of a cell, then if we could quantify these and scale them up, looking for those patterns should be really useful.
Where did you take it from there?
We devised Cell Painting to help pack as much information as possible into a single assay, instead of relying on whatever the biologist decided to specifically stain for. The Cell Painting assay uses six fluorescent dyes to reveal eight cellular components or organelles: the nucleus, the nucleoli, cytoplasmic RNA, the endoplasmic reticulum, the mitochondria, the plasma (cell) membrane, the Golgi complex and the F-actin cytoskeleton. This is like a hit list of microscopists’ favorite dyes because they show parts of the cell that respond to all kinds of stressors, like drugs or genetic mutations.
Still, I didn’t expect that image-based assays could be as powerful as profiling based on RNA transcripts or proteins. In a single experiment, you can measure thousands of transcripts or hundreds of proteins. Yet we only have a handful of stains for a given image. I thought, how far can you get?
I lost a lot of sleep in the early days, trying to rule out artifacts and improve the method and see if it would really be worthwhile. But then the next decade or so brought discovery after discovery based on using images in a profiling way.
Today, machine learning can extract a lot of information from images. Were these algorithms part of the original version of CellProfiler that launched in 2005?
Not at all. CellProfiler’s function was to turn images into numbers by letting classical image processing algorithms measure the images’ properties. It wasn’t until later that machine learning came into play in three ways.
First, machine learning can find the borders of cells and other subcellular structures. Deep learning algorithms are now more accurate but also often easier for biologists to apply — it’s the best of both worlds.
Second, let’s say CellProfiler extracts a thousand features per cell. If you want to know if cells are metastatic, and if that’s a phenotype you can recognize by eye, you can use supervised machine learning to teach the computer what metastatic cells and nonmetastatic cells look like based on those features.
A third way is a very recent development. Rather than using CellProfiler to identify cells and then extract their features, you just give the entire image in all of its raw pixel glory to a deep learning neural network, and it will extract all kinds of features that don’t necessarily map very well to a biologist’s preconceived ideas about relevant features, like cell size or what might stain red in the nucleus. We are finding this kind of feature extraction to be quite powerful.
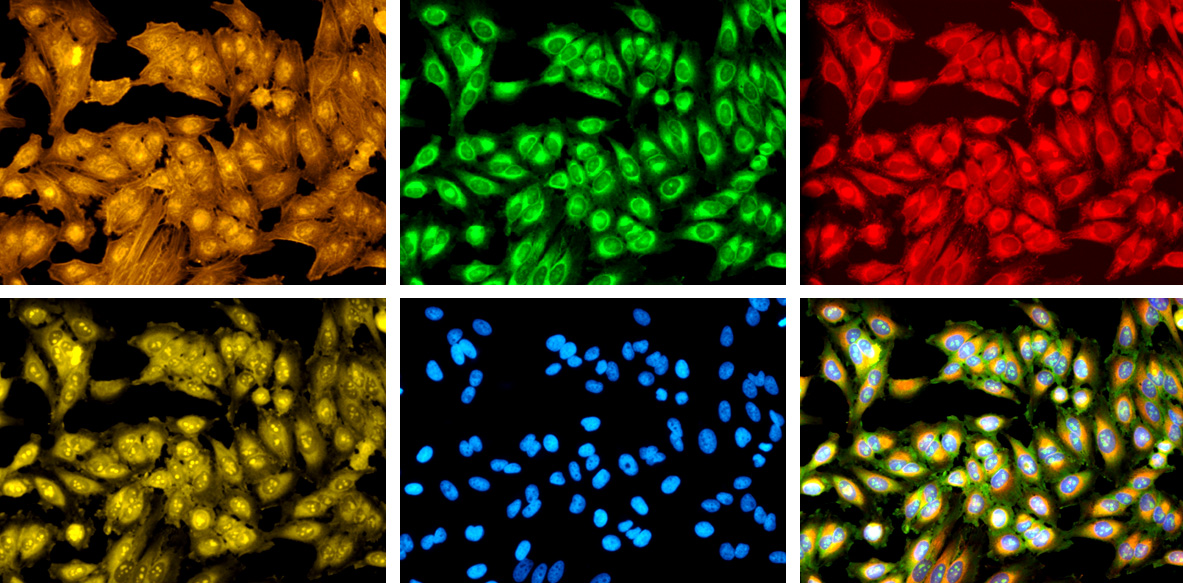
In each of these images, the cells have been treated with one or more dyes that stain specific cellular features. By registering the precise positions of more than a thousand of these features, CellProfiler and other tools can identify the types of individual cells and pathological states that they may be displaying.
Broad Institute
How did you equip yourself in machine learning, a field that can seem pretty foreign and daunting to biologists?
If you had told college-aged Anne, “22 years from now, you’re going to be leading a research group focused on AI,” I would have said you’re insane. It would not have been possible to make this shift into machine learning without having made friends with machine learning experts — particularly Jones.
After he and I finished our training at MIT, we started a lab together at the Broad Institute in 2007, and we brainstormed a lot about how machine learning could help biologists. What allowed these ideas to percolate and develop was both of us hopping over the fence and getting familiar with the terminology and power of both sides, biology and computer science. It’s really a productive partnership.
And it’s not just Jones anymore. My group is about 50-50 in terms of people coming from the biology side versus the computational side.
You’ve had a lot of success in promoting interdisciplinary work.
I like bringing people together. My lab welcomes people who are curious and have different ideas — kind of the opposite of the toxic tech bro culture where it’s “we’re important, we do our thing, and don’t ask a question unless you want to get mocked.” When I realized it’s hard to be a woman in computer science, I realized immediately that it’s much harder to be in a racial minority in science in general.
We focus on whether the person has skills and interests that complement the group, whether they are curious about areas outside their domain, and whether they can communicate well to people without the same training. And without explicitly trying, my lab has been much more diverse than average for a computational lab at a top-tier institution. And the majority of the independent labs launched from among my alumni are led by women or people from minoritized groups.
I wonder how many people don’t think they’re racist or sexist, but when hiring they are, like, “This guy talks like me, he understands our language and jargon, he understands our domain,” not to mention “he’s the kind of person I’d like to have a beer with.” You can see how that would end up with a group that is homogeneous in demographics but also in domain expertise and experience.
These days, your group focuses on developing image-based profiling tools to accelerate drug discovery. Why did you choose that?
Several lines of evidence helped solidify that mission. One came from head-to-head experiments in 2014 that showed image-based profiles could be just as powerful as transcriptional profiles.
Another was described in our 2017 eLife paper, where we overexpressed a couple hundred genes in cells and found that half of them had an impact on cell morphology. By grouping the genes based on the imaging data, you can see in one beautiful cluster analysis what has taken biologists decades to piece together about various signaling pathways: over here, all the genes related to the RAS pathway involved in cancer; over there, the genes in the Hippo pathway that regulates tissue growth, and so on.
Looking at that visualization and realizing we had reconstituted a lot of biological knowledge for this set of genes in a single experiment — maybe a couple of weeks’ work — was really remarkable to me. It made us decide to invest more time and energy into developing this research trajectory.
In a 2018 Cell Chemical Biology paper, Janssen Pharmaceutica researchers dug up images sitting around from old experiments — where they had measured only the one thing they had cared about — and found that there was often enough information in those images to predict results from other assays the company conducted. About 37% of assay results could be predicted by machine learning using images they had lying around. This really got the attention of big pharma! Replacing a large-scale drug assay with a computational query saves millions of dollars each time.
In a consortium I helped launch in 2019, a dozen companies and nonprofit partners are working to create a massive Cell Painting data set of cells treated with more than 120,000 compounds and subjected to 20,000 genetic perturbations. The goal is to speed drug discovery by determining the mechanism of action of potential drugs before they go into clinical trials.
What are some examples of how image-based profiling can help find new drugs?
Recursion Pharmaceuticals is the company farthest along in using image-based profiling, with four drug compounds going into clinical trials. I serve on their scientific advisory board. Their basic approach is to say, let’s perturb a gene known to cause a human disease and see what happens to cells as a result. And if the cells change in any measurable way, can we find a drug that causes the unhealthy-looking cells to go back to looking healthy?
They’ve taken it a step further. Without even testing the drugs on the cells, they can computationally predict which disease phenotypes might be mitigated by which compounds, based on previous tests showing a compound’s impact on cells. I know this strategy works, because my lab has been working on the same thing in a project we just preprinted, though using relatively primitive computational techniques.
I’ve been collaborating with Paul Blainey at MIT and J.T. Neal at the Broad Institute on this genetic bar-coding technique that would let us mix a bunch of genetic perturbations in cells and then use bar-coding to figure out which cell got which genetic reagent. That allows us to mix together 200 normal and 200 mutated human proteins in a single well that we can treat with a drug. For each well, we’re testing whether this drug is useful for any of these 200 diseases. So it’s 200 times cheaper than doing 200 individual drug screens.
We got internal funding to do a pilot with 80 drugs and are seeking funding to test about 6,800 drugs. If we do this well, it may be that about a year from now, the outcome of this experiment suggests actual drugs for these disorders that doctors could prescribe after reading our paper.
What excites you about the future of image-based profiling in biomedical research — and perhaps more broadly, about the future of AI in this realm?
We’re already at the point where implementing existing machine learning methods improves the drug discovery process. But I can see a future, beyond the current capabilities of image-based profiling, where you start gaining exponentially, in leaps and bounds.
All the machine learning algorithms we’re using were developed for social media to identify faces and for financial institutions to identify unusual transactions — that sort of thing. I think putting some more attention toward biological domains and cellular images specifically could really move things forward faster.