Cell-Bacteria Mergers Offer Clues to How Organelles Evolved
The cells of mealybugs contain two types of symbiotic bacteria, one nested inside the other. The lives of these bacteria are so intertwined with their host’s that they function almost like organelles: They draw on their host’s genome to complete the set of enzymes essential to their own metabolic pathways.
Introduction
There are few relationships in nature more intimate than those between cells and the symbiotic bacteria, or endosymbionts, that live inside them. In these partnerships, a host cell typically provides protection to its endosymbiont and gives it a way to propagate, while the endosymbiont provides key nutrients to the host. It’s a deeply cooperative arrangement, in which the genomes of the host and the endosymbiont even seem to contribute complementary pieces to each other’s metabolic and biosynthetic pathways.
The revealed intricacy of these partnerships continues to hold surprises. In a new study appearing today in Cell, scientists show that a complex three-way symbiosis between an insect cell and two species of bacteria — one an endosymbiont of the other — deeply intertwines the organisms’ genomes and physiologies. Those results may illuminate how mitochondria and other organelles arose from ancient endosymbionts in the earliest eukaryotic cells.
When cells need to quickly acquire a new metabolic trait to survive, their best option may be to borrow one from other organisms. Horizontal transfers can move a few genes between cells, but the chances of horizontally acquiring the complete suite of genes for a complex metabolic pathway are vanishingly small. So the easiest solution is often for cells with dissimilar abilities and complementary needs to merge, explains John McCutcheon, an endosymbiosis researcher at the University of Montana. These mergers are not uncommon in nature. Secondary and tertiary mergers are even known to have occurred, producing the cellular equivalent of a set of nested Russian dolls.
One such Russian-doll merger occurred about 100 million years ago, when the small insect pests called mealybugs acquired a bacterial endosymbiont, Tremblaya. Subsequently, Tremblaya acquired several other bacteria including Moranella. Eventually the others were lost and only Moranella remained. It’s not known how long the Moranella endosymbiosis has been going on, but it is probably on the order of tens of millions of years. The result is that the mealybug cell contains a bacterium that contains another bacterium – an arrangement discovered back in 2001 by Carol von Dohlen, a biologist at Utah State University.
In 2011, von Dohlen and McCutcheon published the sequenced genomes of these two bacteria. Each of the genomes had lost genes, but together they had the full complement of genes coding for enzymes in biosynthetic pathways for essential amino acids. Thus, Tremblaya and Moranella work together to produce essential amino acids for themselves as well as the ones that the mealybug cannot find in its strict sap diet.
But the two bacterial genomes were missing other genes as well, and although they complemented one another for amino acid synthesis, they seemed unable to make enzymes crucial to other metabolic pathways. That led McCutcheon to wonder whether the host insect’s genome contained the genes that filled those holes.

The mealybug (Planococcus citri) is a well-known garden pest, widely distributed around the world.
Alex Wild
In a paper published in 2013, McCutcheon and colleagues showed that this was indeed the case. They also noticed that although those genes sat inside the nuclei of the insect host cells, many of them had clearly not started out as mealybug genes because they coded for synthesizing peptidoglycans, the main components of bacterial cell walls. Those genes had to have been horizontally transferred into the mealybug nuclear genome from bacteria.
The genomic evidence therefore suggested, but did not prove, that the Moranella endosymbiont might rely on gene products from the mealybug’s nuclear genome to make its cell walls. If so, however, it meant that the products of the insect’s genes had to move from the host nucleus through five cell membranes (three in Tremblaya and two in Moranella) to reach the inside of the most deeply nested bacteria, where the peptidoglycans are made. That unproved proposition seemed highly unlikely.
In addition, it would mean that genes from at least three disparate sources — authentic mealybug nuclear genes, various bacterial genes acquired by the mealybug nuclear genome, and Moranella genes — were all working together in a complex biosynthetic pathway. That also seemed unlikely.
The hypothesis taking shape looked ungainly, even to the researchers. “It’s so complicated, and for all this stuff to work together, it’s almost just ridiculous,” McCutcheon said. Still, they had enough confidence in their genomic data to devise a way to test it. “We wanted to figure out a pathway where we could go in and actually prove that what we were seeing in the genomics actually worked the way we thought it would.”
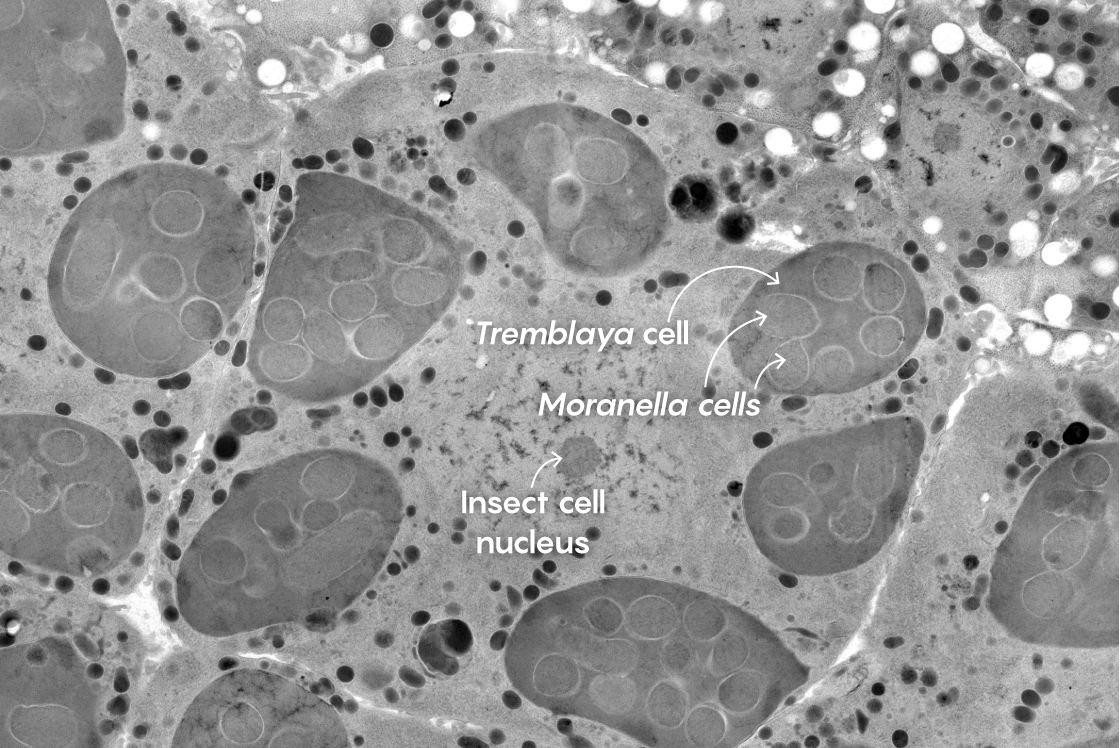
This electron micrograph shows a close-up of a mealybug cell at center. Within it are six darker cells of the bacterium Tremblaya. Within each of the Tremblaya cells, several cells of Moranella can be seen. Surprisingly, some products of the mealybug nucleus seem to penetrate all the way into the Moranella cells.
Mark Ladinsky, California Institute of Technology
Symbionts and Metabolism
In a paper published today in Cell, McCutcheon, DeAnna Bublitz (a senior scientist in McCutcheon’s laboratory) and their colleagues describe the clever trick that enabled them to accomplish this. They exploited a unique feature of peptidoglycans: that they are made with D-alanine, an amino acid found nowhere else in cell metabolism. In the experiments, the researchers gave growing cultures of mealybug cells versions of D-alanine that were tagged with either a heavy nitrogen isotope or fluorescent compounds, which allowed them to trace its location in the cell and its various metabolic transformations.
Their findings confirmed that the complete biochemistry of peptidoglycan synthesis was occurring inside the nested Moranella endosymbionts.
When the researchers first saw the results of the heavy-isotope experiment, Bublitz recalls, she was in a basement laboratory with other grad students and research scientists. “The screen came up and started to show that the pattern we expected with peptidoglycan was real, but we took a moment to go stand back in the hallway and look at the screen from very, very far away to make sure that what we were seeing was still visible,” she said. “It was that level of disbelief.” The researchers even called in strangers and asked them to describe the image on the screen to see “if it was as obvious to them as blind reviewers as it was to us, and that we weren’t just seeing what we wanted to see.”
“The thing that makes me most excited is that this shows that these complicated endosymbioses work,” McCutcheon said. He was also struck that Moranella and Tremblaya are so integrated into the mealybug cells that they are effectively part of them: “Seeing the genetic complexity that underlies it really erodes all functional distinction between endosymbiont and organelle.”
W. Ford Doolittle, an evolutionary and molecular biologist at Dalhousie University in Nova Scotia, says this study represents “a necessary and exciting amount of ground-truthing” because it tested out the biochemistry implied by the genomic data and showed that the cells are actually making what they’re supposed to be making. “I do think this is a very significant paper,” he said.
“It’s quite a tour de force to be able to actually show that the innermost bacterium makes the cell walls,” said Seemay Chou, a biochemist studying interactions between animals and microbes at the University of California, San Francisco. “It’s a very technically challenging thing to demonstrate.”
“What is super wild about this is that the division of labor requires that the host transport something across five different lipid membranes to get to the inner endosymbiont,” she added.
It’s unclear at this point whether the symbiotic cells are using the same molecular mechanisms that eukaryotic cells normally use to shuttle proteins across cell membranes or whether they had to invent new ways to move this cargo around. According to McCutcheon, some evidence indicates that Tremblaya, the middleman in this three-way symbiosis, actively transports the gene products between the host nucleus and the Moranella genome, because none of the heavy isotope-tagged metabolites were ending up in Tremblaya. “It is participating somehow, but that ‘how’ is really quite a mystery,” he said.
The Controlling Partner
There is at least one other known example of an endosymbiosis that involves shared genes. Paulinella, a protist, evolved a “second chloroplast” or chromatophore about 100 million years ago by acquiring a cyanobacterial endosymbiont. The chromatophore genome works in complement with the host protist genome to make the chromatophore’s peptidoglycan layer.
McCutcheon speculated that in both Paulinella and the mealybug-Tremblaya-Moranella endosymbiosis, the genomic mosaic for making peptidoglycans may enable the eukaryotic host to control its bacterial endosymbionts. If the endosymbiont replicated too quickly, it could kill the host; by limiting the rate at which the bacteria can build cell walls, the host keeps its residents in check. Why Moranella continues to make a cell wall remains a mystery, however, because it is already safely encased inside both Tremblaya and the host cell. “Clearly, the fact that it has to do all this suggests it’s important for some reason,” Chou said.
McCutcheon thinks that the mealybug ménage à trois may hold clues about the evolution of the very oldest, and probably best-known, organelle: mitochondria.
Mitochondria evolved from an alphaproteobacteria that was engulfed by a prokaryote (most likely a member of Archaea) between 1.5 billion and 2 billion years ago. Because most living bacteria have peptidoglycan cell walls, it is likely that ancient ones — including the ancestor of mitochondria — did too. Bublitz hypothesizes that the alphaproteobacteria got taken up by another cell or actively invaded it, and that the host cell was able to co-opt the peptidoglycan pathway and control its endosymbiont’s replication. It’s possible the host and the early mitochondria eventually became further integrated, so much so that the host no longer needed the peptidoglycan pathway to control the mitochondria’s division. Ceding control over peptidoglycan synthesis might be “one of those first steps in transitioning from an autonomous bacterium to some kind of functional organelle,” Bublitz speculated.
Because mitochondria are so ancient, and because they evolved only once, it is difficult to reconstruct exactly how that endosymbiosis evolved. What we know is that over time mitochondrial genomes lost genes, some of which came to be inserted into the nuclear genome. Today, in most animals complicated enough to have bilateral symmetry, the mitochondrial genomes retain only 37 genes; the mitochondria rely on more than 1,000 genes now in the nucleus to function. (In contrast, eukaryotic microbes ordinarily have between three and 69 mitochondrial genes.) But a little-known fact is that many of those nuclear genes didn’t come from the ancient mitochondrial genome; they came from other bacteria in horizontal transfer events, McCutcheon says.
The origin of these horizontally transferred genes is a hotly debated issue in mitochondrial biology. One possibility is that the alphaproteobacteria acquired them from other bacteria before the endosymbiotic event that produced the eukaryotic cell, and the genes subsequently moved over to the host nucleus. Another is that the horizontally transferred genes arrived in the nuclear genome directly from various bacteria over time, after the eukaryotic cell evolved.
Although we can’t be completely certain how the oldest organelle evolved, the mealybug-Tremblaya-Moranella symbiosis demonstrates that the second evolutionary scenario can work — that genes from different bacterial infections can accumulate slowly and become integrated into functional pathways in a single entity.
What makes mitochondria unique among endosymbionts-turned-organelles is that they are the oldest example, McCutcheon said. “But their antiquity makes them hard to study, and it makes it hard to infer what happened before they became organelles. I think the thing about Paulinellais that it gives us a window into what might’ve happened.”
Although mitochondria have been uniquely successful, not all endosymbioses end as happily. Earlier work by McCutcheon showed that cicadas have a bacterial endosymbiont, Hodgkinia cicadicola, that fragmented into more than two dozen lineages inside the cicada cells. Those lineages each contain only varying subsets of the Hodgkinia genome. Together, all of the lineages have the full complement of genes needed to make the essential amino acids the cicadas depend on. But McCutcheon thinks this is an example of a nonadaptive endosymbiont evolution that creates difficulties for the host: The cicada eggs need to pick up a full complement of the endosymbionts to survive, and the variability in what different combinations of endosymbionts provide poses problems.
McCutcheon and Bublitz are now working to figure out why some endosymbioses evolve into stable, successful partnerships while others spiral out of control or degrade. “Right now, there’s no smoking gun as to what allows one to persist,” Bublitz said.