DNA Has Four Bases. Some Viruses Swap in a Fifth.
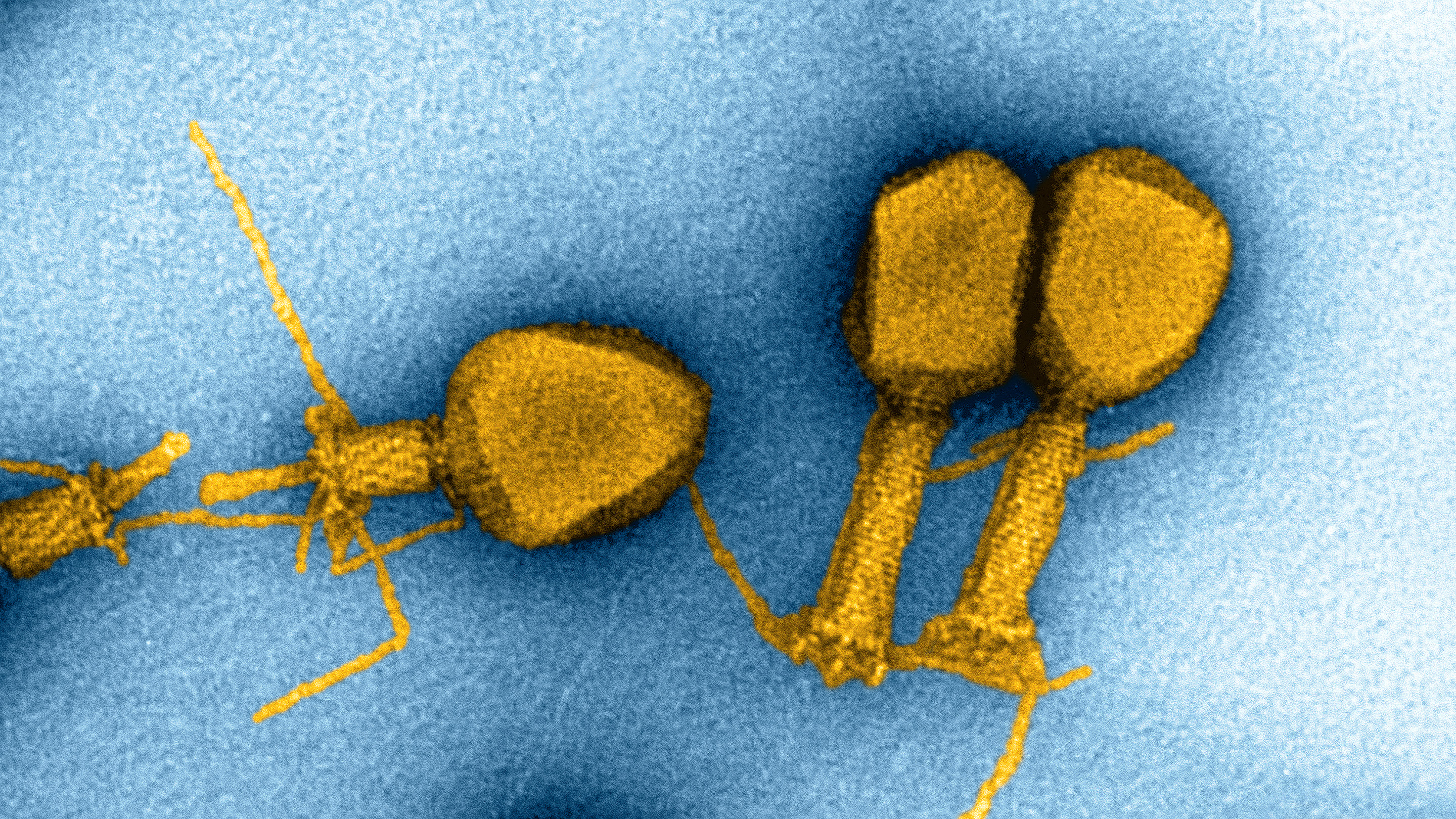
Some viruses replace one of the familiar A, C, T or G nucleotide bases in their DNA with a modified fifth base. Scientists are exploring how widespread these substitutions might be.
Omikron
Introduction
All life on Earth rests on the same foundation: a four-letter genetic alphabet spelling out a repertoire of three-letter words that specify 20 amino acids. These basic building blocks — the components of DNA and their molecular interpreters — lie at biology’s core. “It’s hard to imagine something more fundamental,” said Floyd Romesberg, a synthetic biologist at the pharmaceutical company Sanofi.
Yet life’s foundational biochemistry can be full of surprises. A few decades ago, researchers found viruses that had swapped one of the four bases in their DNA for a novel fifth one. Now, in a trio of papers published in Science in April, three teams have identified dozens of other viruses that make this substitution, as well as the mechanisms that make it possible. The discoveries raise the thought-provoking possibility that this kind of fundamental genomic change could be much more widespread and important in biology than anyone imagined.
“Here was this wonderful validation that right under our noses, nature has been expanding,” said Stephen Freeland, a biologist at the University of Maryland, Baltimore County.
“It really speaks to the adaptability of the genetic alphabet,” Romesberg said.
Researchers have long been intrigued by the possibility that evolution could have gone in a different direction with DNA’s four bases: adenine (A), thymine (T), cytosine (C) and guanine (G). Perhaps there could have been more than four of them, or they could have had very different chemical or binding properties, or they could have used a different set of rules to represent information. Synthetic biologists like Romesberg have explored this by engineering artificial base pairs and additional amino acids to produce novel proteins. Even so, because an organism’s survival depends on keeping its genetic alphabet and code intact, the precise ingredients in DNA’s recipe are thought to have been largely locked in by evolution for billions of years — making them “frozen accidents,” in the words of Francis Crick.
But some exceptions have cropped up. In 1977, for instance, researchers in the Soviet Union found something peculiar while looking at a virus that infects photosynthetic bacteria: All the A’s in the genome had been replaced with an alternative base, 2-aminoadenine, which was later dubbed Z. Usually, C pairs with G and T pairs with A to form double-stranded DNA. But in this virus, with no A’s to be found, T paired with Z. (During gene transcription, T-Z was still treated as though it were T-A.)
The Z base looks like a chemical modification of A; it’s an adenine nucleotide with an extra attachment. But that modest change allows Z to form a triple hydrogen bond with T, which is more stable than the double bond that holds together A-T.
The finding was intriguing but seemed like an isolated case. “It came as a kind of curiosity, something really weird and of no general significance,” said Philippe Marlière, a geneticist at the University of Evry in France and one of the leaders of the new research on Z genomes. “And so it settled into oblivion, more or less.”
But since the alterations were “at the deepest level of chemical organization,” he said, “my instinct told me this is not just an anecdote. This is a profound violation.”
In the early 2000s, Marlière and his colleagues sequenced the genome of the bacteriophage that the Russian team had studied, and they pinpointed a genetic sequence associated with production of the Z base. For the next 15 years, they searched for matches in databases of other viral genomes. Another group, led by researchers in Illinois and China, independently joined the effort.
The scientists have now reported finding the Z substitution in more than 200 phages. Further analysis of the viral genomes allowed the research groups to uncover a key enzyme for making Z, as well as an enzyme that degrades free-floating A nucleotides, making Z more likely to be taken up during DNA synthesis.
Samuel Velasco/Quanta Magazine; source: doi.org/10.1038/d41586-021-01157-x
But the biggest surprise was that the viruses had a polymerase enzyme dedicated to pairing Z bases with T’s during DNA replication. “It was like a fairy tale,” said Marlière, who had been hoping to find such a polymerase. “Our wildest dreams came true.”
That’s because while scientists have uncovered other examples of bacteriophages making nucleotide substitutions, this “is the first polymerase that is really shown to selectively exclude a canonical nucleotide,” said Peter Weigele, a researcher at New England Biolabs who studies the biosynthesis of noncanonical bases. The system evolved to allow “a reprogramming,” Romesberg said — one that could potentially provide new insights into how polymerases function, and how to engineer them.
Z and other modified DNA bases seem to have evolved to help viruses evade the defenses with which bacteria degrade foreign genetic material. The eternal arms race between bacteriophages and their host cells probably provides enough selection pressure to affect something as seemingly “sacrosanct” as DNA, according to Romesberg. “Right now, everyone thinks the modifications are just protecting the DNA,” he said. “People almost trivialize it.”
But something more may be at work: The triple bond of Z, for instance, might add to DNA’s stability and rigidity, and perhaps influence some of its other physical properties. Those changes could carry advantages beyond hiding from bacterial defenses and could make such modifications more broadly significant.
After all, no one really knows how many viruses may have played with their DNA like this. “Standard [genome sequencing] methods for looking for biological diversity in nature would fail to find these,” said Steven Benner, a chemist at the Foundation for Applied Molecular Evolution in Florida who has synthesized several artificial base pairs, “because we are looking in a way that assumes a common biochemistry that is not present.”
These kinds of overlooked substitutions might even turn up in more than viruses. “Maybe we missed some of this in the bacterial world, right?” said Chuan He, a chemical biologist at the University of Chicago.
Synthetic biology has (again) shown that this is possible. For years, Marlière’s team has been evolving E. coli that use a modified base instead of T nucleotides. Huimin Zhao, a chemist at the University of Illinois, Urbana-Champaign and a leader of some of the recent Z genome work, is trying to get E. coli and potentially other cells to incorporate Z as the viruses do.
Romesberg thinks that these findings could raise questions about modifications of bacterial DNA that were thought to be epigenetic — that is, changes made to nucleotides after the DNA was synthesized, usually to influence gene expression. The Z substitution, he said, “shows that things that you might have thought were epigenetic might not be.”
“I think people need to look under rocks that were thought to be understood,” he added. “That’s where surprises come from.”
But there’s also plenty of room for surprises in less well-studied places, because “we can’t cultivate most of Earth’s microbes,” said Carol Cleland, a philosopher of science at the University of Colorado, Boulder. “Is there other stuff out there that we just aren’t able to recognize?”
Marlière wonders, for example, if scientists might one day stumble on more than one kind of base modification in a single genome. Or perhaps they’ll find a change to the molecular backbone of DNA, in which case “it would no longer be DNA,” he said. “It would be something else.”
We need to “stop taking the components of molecular biology as we know them for granted,” Freeland said. “Purely because our instrumentation has gotten better and we’ve looked harder, everything that we thought was standard and universal is just falling away.”






