DNA’s Histone Spools Hint at How Complex Cells Evolved

By managing gene expression in complex cells, octets of histone proteins helped to enable the explosive diversity of eukaryotic life.
Jason Lyon for Quanta Magazine
Introduction
Molecular biology has something in common with kite-flying competitions. At the latter, all eyes are on the colorful, elaborate, wildly kinetic constructions darting through the sky. Nobody looks at the humble reels or spools on which the kite strings are wound, even though the aerial performances depend on how skillfully those reels are handled. In the biology of complex cells, or eukaryotes, the ballet of molecules that transcribe and translate genomic DNA into proteins holds centerstage, but that dance would be impossible without the underappreciated work of histone proteins gathering up the DNA into neat bundles and unpacking just enough of it when needed.
Histones, as linchpins of the apparatus for gene regulation, play a role in almost every function of eukaryotic cells. “In order to get complex, you have to have genome complexity, and evolve new gene families, and you have to have a cell cycle,” explained William Martin, an evolutionary biologist and biochemist at Heinrich Heine University in Germany. “And what’s in the middle of all this? Managing your DNA.”
New work on the structure and function of histones in ancient, simple cells has now made the longstanding, central importance of these proteins to gene regulation even clearer. Billions of years ago, the cells called archaea were already using histones much like our own to manage their DNA — but they did so with looser rules and much more variety. From those similarities and differences, researchers are gleaning new insights, not only into how the histones helped to shape the origins of complex life, but also into how variants of histones affect our own health today. At the same time, though, new studies of histones in an unusual group of viruses are complicating the answers about where our histones really came from.
Dealing With Too Much DNA
Eukaryotes arose about 2 billion years ago, when a bacterium that could metabolize oxygen for energy took up residence inside an archaeal cell. That symbiotic partnership was revolutionary because energy production from that proto-mitochondrion suddenly made expressing genes much more metabolically affordable, Martin argues. The new eukaryotes suddenly had free rein to expand the size and diversity of their genomes and to conduct myriad evolutionary experiments, laying the foundation for the countless eukaryotic innovations seen in life today. “Eukaryotes are an archaeal genetic apparatus that survives with the help of bacterial energy metabolism,” Martin said.
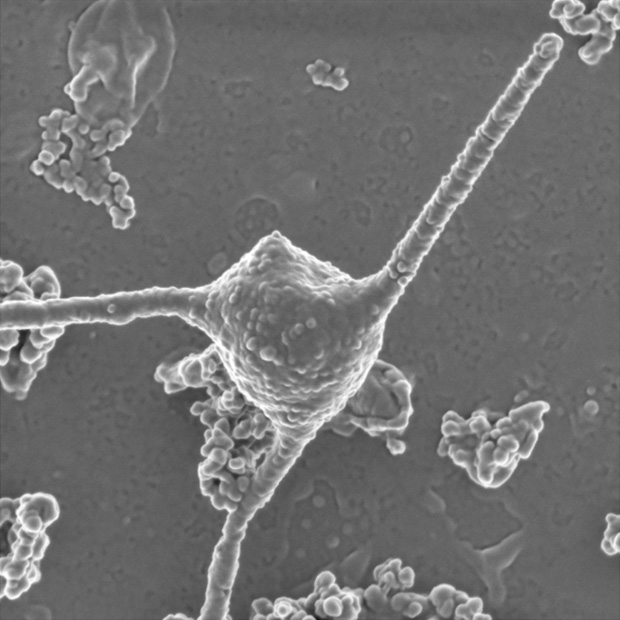
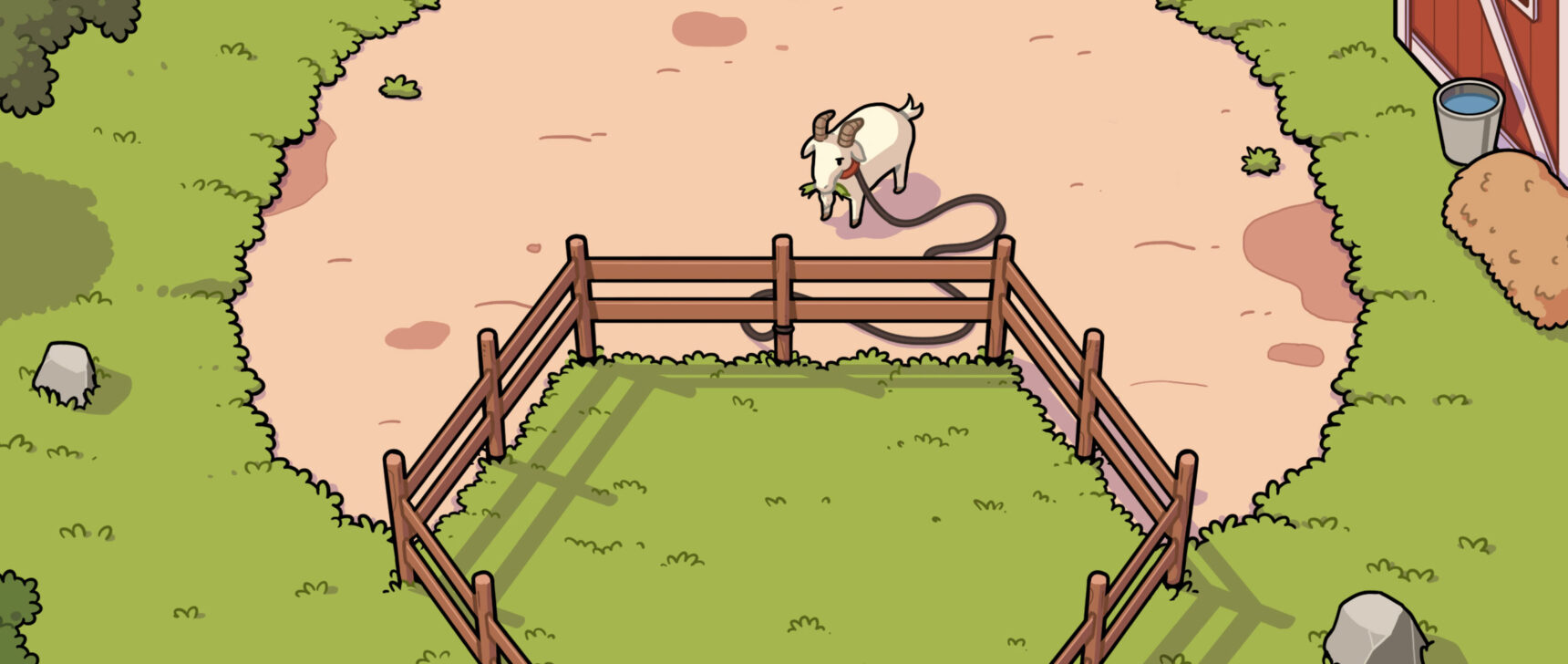
Eukaryotes arose about 2 billion years ago through a partnership between two simple prokaryotes. An archaeal cell (top) became the permanent host of endosymbiotic bacteria, which evolved into energy-producing mitochondria.
Eukaryotes arose about 2 billion years ago through a partnership between two simple prokaryotes. An archaeal cell (left) became the permanent host of endosymbiotic bacteria (right), which evolved into energy-producing mitochondria.
Imachi H, Nobu MK and JAMSTEC; DENNIS KUNKEL MICROSCOPY / Science Source
But the early eukaryotes went through serious growing pains as their genomes expanded: The larger genome brought new problems stemming from the need to manage an increasingly unwieldy string of DNA. That DNA had to be accessible to the cell’s machinery for transcribing and replicating it without getting tangled up in a hopeless spaghetti ball.
The DNA also sometimes needed to be compact, both to help regulate transcription and regulation, and to separate the identical copies of DNA during cell division. And one danger of careless compaction is that DNA strands can irreversibly bind together if the backbone of one interacts with the groove of another, rendering the DNA useless.
Bacteria have a solution for this that involves a variety of proteins jointly “supercoiling” the cells’ relatively limited libraries of DNA. But eukaryotes’ DNA management solution is to use histone proteins, which have a unique ability to wrap DNA around themselves rather than just sticking to it. The four primary histones of eukaryotes — H2A, H2B, H3 and H4 — assemble into octamers with two copies of each. These octamers, called nucleosomes, are the basic units of eukaryotic DNA packaging.
By curving the DNA around the nucleosome, the histones prevent it from clumping together and keep it functional. It’s an ingenious solution — but eukaryotes didn’t invent it entirely on their own.
Samuel Velasco/Quanta Magazine
Back in the 1980s, when the cellular and molecular biologist Kathleen Sandman was a postdoc at Ohio State University, she and her adviser, John Reeve, identified and sequenced the first known histones in archaea. They showed how the four principal eukaryotic histones were related to each other and to the archaeal histones. Their work provided the early evidence that in the original endosymbiotic event that led to eukaryotes, the host was likely to have been an archaeal cell.
But it would be a teleological mistake to think that archaeal histones were just waiting for the arrival of eukaryotes and the chance to enlarge their genomes. “A lot of these early hypotheses looked at histones in terms of their ability to allow the cell to expand its genome. But that doesn’t really tell you why they were there in the first place,” said Siavash Kurdistani, a biochemist at the University of California, Los Angeles.
As a first step toward those answers, Sandman joined forces several years ago with the structural biologist Karolin Luger, who solved the structure of the eukaryotic nucleosome in 1997. Together, they worked out the crystallized structure of the archaeal nucleosome, which they published with colleagues in 2017. They found that the archaeal nucleosomes are “uncannily similar” in structure to eukaryotic nucleosomes, Luger said — despite the marked differences in their peptide sequences.
Archaeal nucleosomes had already “figured out how to bind and bend DNA in this beautiful arc,” said Luger, now a Howard Hughes Medical Institute investigator at the University of Colorado, Boulder. But the difference between the eukaryotic and archaeal nucleosomes is that the crystal structure of the archaeal nucleosome seemed to form looser, Slinky-like assemblies of varying sizes.
In a paper in eLife published in March, Luger, her postdoc Samuel Bowerman, and Jeff Wereszczynski of the Illinois Institute of Technology followed up on the 2017 paper. They used cryo-electron microscopy to solve the structure of the archaeal nucleosome in a state more representative of a live cell. Their observations confirmed that the structures of archaeal nucleosomes are less fixed. Eukaryotic nucleosomes are always stably wrapped by about 147 base pairs of DNA, and always consist of just eight histones. (For eukaryotic nucleosomes, “the buck stops at eight,” Luger said.) Their equivalents in archaea wind up between 60 and 600 base pairs. These “archaeasomes” sometimes hold as few as three histone dimers, but the largest ones consist of as many as 15 dimers.
They also found that unlike the tight eukaryotic nucleosomes, the Slinky-like archaeasomes flop open stochastically, like clamshells. The researchers suggested that this arrangement simplifies gene expression for the archaea, because unlike eukaryotes, they don’t need any energetically expensive supplemental proteins to help unwind DNA from the histones to make them available for transcription.
That’s why Tobias Warnecke, who studies archaeal histones at Imperial College London, thinks that “there’s something special that must have happened at the dawn of eukaryotes, where we transition from just having simple histones … to having octameric nucleosomes. And they seem to be doing something qualitatively different.”
What that is, however, is still a mystery. In archaeal species, there are “quite a few that have histones, and there are other species that don’t have histones. And even those that do have histones vary quite a lot,” Warnecke said. Last December, he published a paper showing that there are diverse variants of histone proteins with different functions. The histone-DNA complexes vary in their stability and affinity for DNA. But they are not as stably or regularly organized as eukaryotic nucleosomes.
As puzzling as the diversity of archaeal histones is, it provides an opportunity to understand the different possible ways of building systems of gene expression. That’s something we cannot glean from the relative “boringness” of eukaryotes, Warnecke says: Through understanding the combinatorics of archaeal systems, “we can also figure out what’s special about eukaryotic systems.” The variety of different histone types and configurations in archaea may also help us deduce what they might have been doing before their role in gene regulation solidified.
A Protective Role for Histones
Because archaea are relatively simple prokaryotes with small genomes, “I don’t think that the original role of histones was to control gene expression, or at least not in a manner that we are used to from eukaryotes,” Warnecke said. Instead, he hypothesizes that histones might have protected the genome from damage.
Archaea often live in extreme environments, like hot springs and volcanic vents on the seafloor, characterized by high temperatures, high pressures, high salinity, high acidity or other threats. Stabilizing their DNA with histones may make it harder for the DNA strands to melt in those extreme conditions. Histones also might protect archaea against invaders, such as phages or transposable elements, which would find it harder to integrate into the genome when it’s wrapped around the proteins.
Kurdistani agrees. “If you were studying archaea 2 billion years ago, genome compaction and gene regulation are not the first things that would come to mind when you are thinking about histones,” he said. In fact, he has tentatively speculated about a different kind of chemical protection that histones might have offered the archaea.
Last July, Kurdistani’s team reported that in yeast nucleosomes, there is a catalytic site at the interface of two histone H3 proteins that can bind and electrochemically reduce copper. To unpack the evolutionary significance of this, Kurdistani goes back to the massive increase in oxygen on Earth, the Great Oxidation Event, that occurred around the time that eukaryotes first evolved more than 2 billion years ago. Higher oxygen levels must have caused a global oxidation of metals like copper and iron, which are critical for biochemistry (although toxic in excess). Once oxidized, the metals would have become less available to cells, so any cells that kept the metals in reduced form would have had an advantage.
During the Great Oxidation Event, the ability to reduce copper would have been “an extremely valuable commodity,” Kurdistani said. It might have been particularly attractive to the bacteria that were forerunners of mitochondria, since cytochrome c oxidase, the last enzyme in the chain of reactions that mitochondria use to produce energy, requires copper to function.
Because archaea live in extreme environments, they might have found ways to generate and handle reduced copper without being killed by it long before the Great Oxidation Event. If so, proto-mitochondria might have invaded archaeal hosts to steal their reduced copper, Kurdistani suggests.
The hypothesis is intriguing because it could explain why the eukaryotes appeared when oxygen levels went up in the atmosphere. “There was 1.5 billion years of life before that, and no sign of eukaryotes,” Kurdistani said. “So the idea that oxygen drove the formation of the first eukaryotic cell, to me, should be central to any hypotheses that try to come up with why these features developed.”
Kurdistani’s conjecture also suggests an alternative hypothesis for why eukaryotic genomes got so big. The histones’ copper-reducing activity only occurs at the interface of the two H3 histones inside an assembled nucleosome wrapped with DNA. “I think there’s a distinct possibility that the cell wanted more histones. And the only way to do that was to expand this DNA repertoire,” Kurdistani said. With more DNA, cells could wrap more nucleosomes and enable the histones to reduce more copper, which would support more mitochondrial activity. “It wasn’t just that histones allowed for more DNA, but more DNA allowed for more histones,” he said.
“One of the neat things about this is that copper is very dangerous because it will break DNA,” said Steven Henikoff, a chromatin biologist and HHMI investigator at the Fred Hutchinson Cancer Research Center in Seattle. “Here’s a place where you have the active form of copper being made, and it’s right next to the DNA, but it doesn’t break the DNA because, presumably, it’s in a tightly packaged form,” he said. By wrapping the DNA, the nucleosomes keep the DNA safely out of the way.
The hypothesis potentially explains aspects of how the architecture of the eukaryotic genome evolved, but it has met with some skepticism. The key outstanding question is whether archaeal histones have the same copper-reducing ability that some eukaryotic ones do. Kurdistani is investigating this now.
The bottom line is that we still don’t definitively know what functions histones served in the archaea. But even so, “the fact that you see them conserved over long distances strongly suggests that they are doing something distinct and important,” Warnecke said. “We just need to find out what it is.”
Histones Are Still Evolving
Although the complex eukaryotic histone apparatus has not changed much since its origin about a billion years ago, it hasn’t been totally frozen. In 2018, a team at the Fred Hutchinson Cancer Research Center reported that a set of short histone variants called H2A.B is evolving rapidly. The pace of the changes is a sure sign of an “arms race” between genes vying for control over regulatory resources. It wasn’t initially clear to the researchers what the genetic conflict was about, but through a series of elegant crossbreeding experiments in mice, they eventually showed that the H2A.B variants dictated the survival and growth rate of embryos, as reported in December in PLOS Biology.
The findings suggested that paternal and maternal versions of the histone variants are mediating a conflict over how to allocate resources to the offspring during pregnancy. They are rare examples of parental-effect genes — ones that don’t directly affect the individual carrying them, but instead strongly affect the individual’s offspring.
The H2A.B variants arose with the first mammals, when the evolution of in utero development rewrote the “contract” for parental investment. Mothers had always invested a lot of resources in their eggs, but mammalian mothers also suddenly became responsible for the early development of their progeny. That set up a conflict: Paternal genes in the embryo had nothing to lose by demanding resources aggressively, while the maternal genes benefited from moderating the burden to spare the mother and let her live to breed another day.
“That negotiation is still ongoing,” said Harmit Malik, an HHMI investigator at the Fred Hutchinson Cancer Research Center who studies genetic conflicts. Exactly how the histones affect the growth and viability of offspring is still not completely understood, but Antoine Molaro, the postdoctoral fellow who led the work and who now leads his own research group at the University of Clermont Auvergne in France, is investigating it.
Some histone variants may cause health problems, too. In January, Molaro, Malik, Henikoff and their colleagues reported that short H2A histone variants are implicated in some cancers: More than half of diffuse large B cell lymphomas carry mutations in them. Other histone variants are associated with neurodegenerative diseases.
But little is yet understood about how a single copy of a histone variant can produce such dramatic disease effects. The obvious hypothesis is that the variants affect the stability of nucleosomes and disrupt their signaling functions, changing gene expression in a way that alters cell physiology. But if histones can act as enzymes, then Kurdistani suggests another possibility: The variants may alter enzymatic activity inside cells.
An Alternative Viral Origin?
Despite the decades-old evidence from Sandman and others that eukaryotic histones evolved from archaeal histones, some intriguing recent work has unexpectedly opened the door to an alternative theory about their origins. According to a paper published on April 29 in Nature Structural & Molecular Biology, giant viruses of the Marseilleviridae family have viral histones that are recognizably related to the four main eukaryotic histones. The only difference is that in the viral versions, the histones that routinely pair up within the octamer (H2A with H2B, and H3 with H4) in eukaryotes are already fused into doublets. The fused viral histones form structures that are “virtually identical to canonical eukaryotic nucleosomes,” according to the paper’s authors.
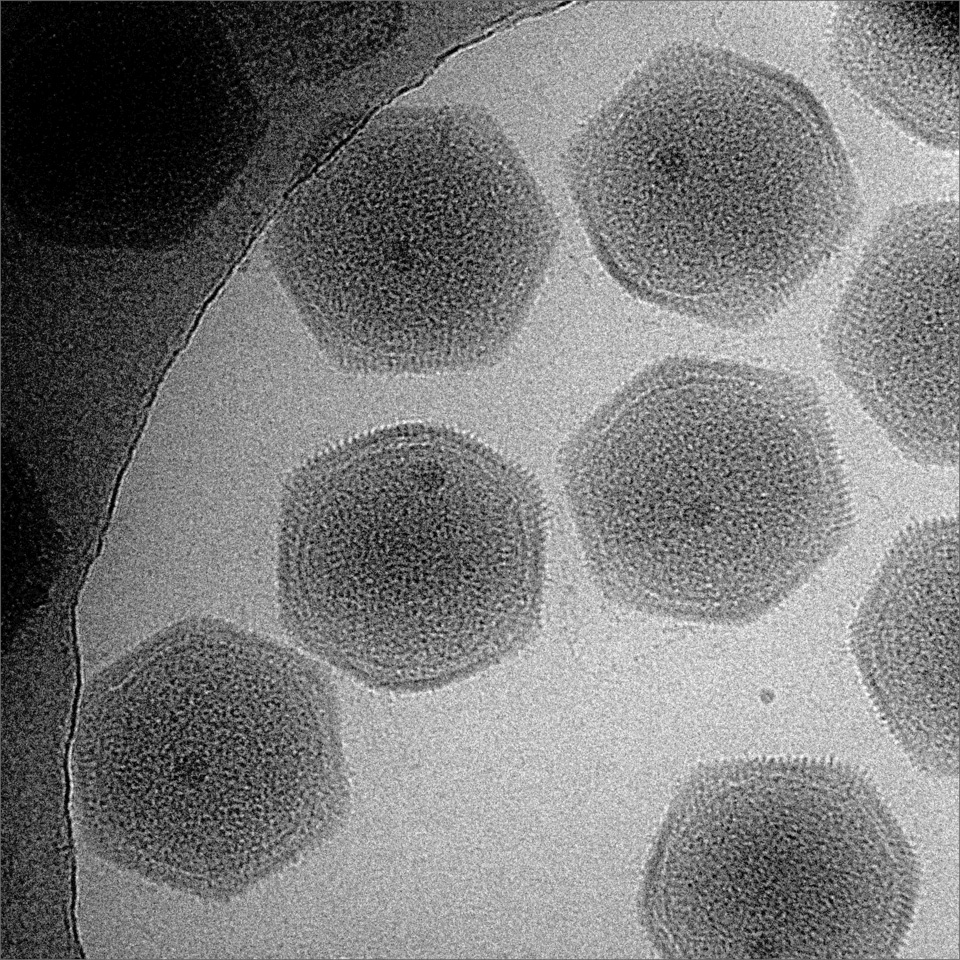
Giant viruses of the Marseilleviridae family were recently found to encode histones that are surprisingly similar to those of eukaryotes.
Courtesy of Kazuyoshi Murata and Kenta Okamoto
Luger’s team posted a preprint on biorxiv.org about viral histones the same day, showing that in the cytoplasm of infected cells, viral histones stay near the “factories” that produce new viral particles.
“Here’s the thing that is really compelling,” said Henikoff, who was among the authors on the new Nature Structural & Molecular Biology paper. “All of the histone variants turn out to be derived from a common ancestor that was shared between eukaryotes and giant viruses. By standard phylogenetic criteria, these are a sister group to eukaryotes.”
It makes a compelling case that this common ancestor is where the eukaryotic histones came from, he says. A “proto-eukaryote” that had histone doublets might have been ancestral to both the giant viruses and eukaryotes and could have passed the proteins along to both lines of organisms a very long time ago.
Warnecke, however, is skeptical about inferring phylogenetic relationships from viral sequences, which are notoriously mutable. As he explained in an email to Quanta, reasons other than shared ancestry might explain how the histones ended up in both lineages. In addition, the idea would require that the histone doublets later “unfused” into the H2A, H2B, H3 and H4 histones, because there are no doublets of those histones in extant eukaryotes. “How and why that would have happened is unclear,” he wrote.
Although Warnecke is not convinced that the viral histones tell us much about the origin of eukaryotic histones, he is fascinated by their possible functions. One possibility is that they help to compact the viral DNA; another idea is that they could be disguising the viral DNA from the host’s defenses.
Histones have had myriad roles since the dawn of time. But it was really in the eukaryotes that they became the linchpins for complex life and countless evolutionary innovations. That’s why Martin calls the histone “a basic building block that never could realize its full potential without the help of mitochondria.”