His Artificial Intelligence Sees Inside Living Cells

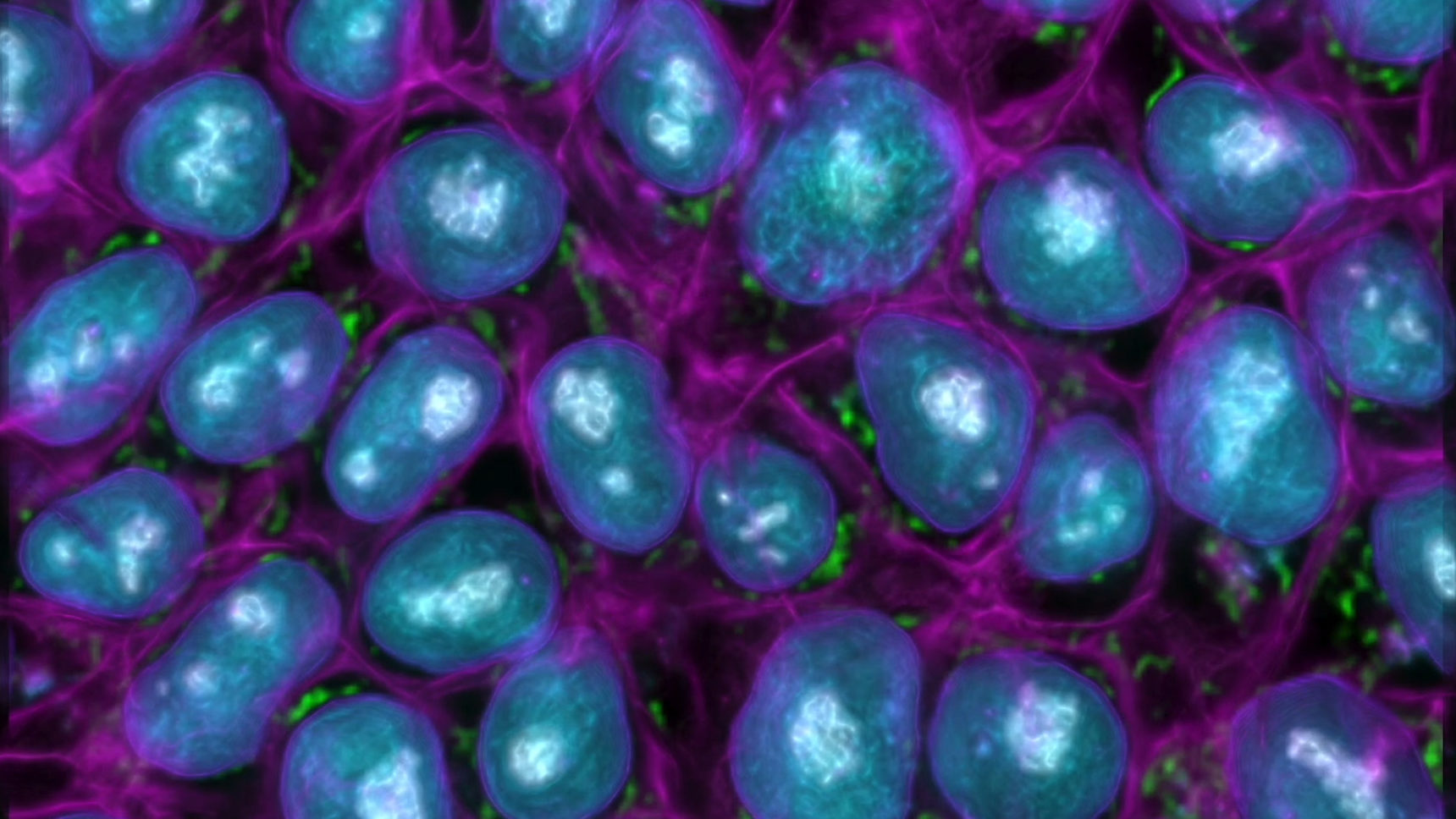
Greg Johnson, a computer vision researcher at the Allen Institute for Cell Science, has shown that deep learning neural networks can pull out details of cellular anatomy from unlabeled micrographs and create sophisticated cell models from them.
Chona Kasinger for Quanta Magazine
Introduction
Your high school biology textbook was wrong about cells. The prototypical human cell — say, a pluripotent stem cell, capable of differentiating into anything from muscle to nerve to skin — isn’t a neat translucent sphere. Nor are its internal parts sitting still and conveniently far apart, like chunks of pineapple suspended in gelatin. In reality, a cell looks more like a pound of half-melted jelly beans stuffed into a too-small sandwich bag. And its contents are all constantly moving, following a choreography more precise and complex than that of a computer chip.
In short, understanding what cells look like on the inside — much less the myriad interactions among their parts — is hard even in the 21st century. “Think of a cell as a sophisticated machine like a car — except every 24 hours, you’ll have two cars in your driveway, and then four cars in your driveway,” said Greg Johnson, a computer vision and machine learning researcher at the Allen Institute for Cell Science. “If you found the smartest engineers in the world and said, ‘Make me a machine that does that,’ they would be totally stumped. That’s what I think of when I think of how little we know about how cells work.”
To view the inner workings of living cells, biologists currently use a combination of genetic engineering and advanced optical microscopy. (Electron microscopes can image cell interiors in great detail, but not with live samples.) Typically, a cell is genetically modified to produce a fluorescent protein that attaches itself to specific subcellular structures, like mitochondria or microtubules. The fluorescent protein glows when the cell is illuminated by a certain wavelength of light, which visually labels the associated structure. However, this technique is expensive and time-consuming, and it allows only a few structural features of the cell to be observed at a time.
But with his background in software engineering, Johnson wondered: What if researchers could teach artificial intelligence to recognize the interior features of cells and label them automatically? In 2018, he and his collaborators at the Allen Institute did just that. Using fluorescence imaging samples, they trained a deep learning system to recognize over a dozen kinds of subcellular structures, until it could spot them in cells that the software hadn’t seen before. Even better, once trained, Johnson’s system also worked with “brightfield images” of cells — images easily obtained with ordinary light microscopes through a process “like shining a flashlight through the cells,” he said.
Instead of performing expensive fluorescence imaging experiments, scientists can use this “label-free determination” to efficiently assemble high-fidelity, three-dimensional movies of the interiors of living cells.
The data can also be used to build a biologically accurate model of an idealized cell — something like the neatly labeled diagram in a high school textbook but with greater scientific accuracy. That’s the goal of the institute’s project.
“We want to be able to say, here’s an average cell, and look at that and dissect it and play with it,” Johnson said. “But because it’s based on data, it would also include all the variation that we would expect. You could say, ‘Let’s look at this [version of the] cell that’s an outlier,’ and ask how it’s organized.”
Johnson’s use of machine learning to visualize cell interiors began in 2010 at Carnegie Mellon University, just before a series of breakthroughs in deep learning technology began to transform the field of artificial intelligence. Nearly a decade later, Johnson thinks that his AI-augmented approach to live cell imaging could lead to software models that are so accurate, they reduce or even altogether eliminate the need for certain experiments. “We want to be able to take the cheapest image [of a cell] that we can and predict from that as much about that cell as we possibly can,” he said. “How is it organized? What’s the gene expression? What are its neighbors doing? For me, [label-free determination] is just a prototype for much more sophisticated things to come.”
Quanta spoke to Johnson about the challenges of basic cell biology and the future of AI in microscopy. The interview has been condensed and edited for clarity.
What’s so hard about seeing inside a living cell?
If you want to look at a cell when it’s alive, there are basically two limitations. We can blast the cell with laser light to get these [fluorescent protein] labels to illuminate. But that laser light is phototoxic — the cell is just basically baking in the sun in the desert.
The other limitation is that these labels are attached to an original protein in the cell that needs to go somewhere and do things. But the protein now has this big stupid fluorescent molecule attached to it. That might change the way the cell works if I have too many labels. Sometimes when you try to introduce these fluorescent labels, your experiment just doesn’t work out. Sometimes, the labels are lethal to the cell.
But when it works, isn’t it good enough? It’s gotten us this far.
If we go back to the cell-as-car metaphor, it’s like you had a car made entirely of glass. You can see things inside the car, but you can’t really tell what you’re seeing or where things are in relation to one another. Then you use this fluorescent molecule as a label to highlight one or two parts in the car. Now you can see the door handles, or you can see however many tires the car has. But sometimes you discover that your “car” has only two wheels, and it doesn’t have any door handles. You say, “I have no idea what this thing is.” It turns out it’s a motorcycle, and we hadn’t even known what motorcycles were because we’d only seen cells with four wheels and door handles, so to speak.
If we could do live cell imaging where we could see everything at the same time, the biological universe would be a very different place. I could take apart the car, look at the car in X-ray vision, and watch cars drive around. Maybe I would be able to build an engine myself. We would at least have a better idea of what the heck is going on.
What inspired you to use deep learning to label what’s inside a cell?
When I saw demonstrations of people generating realistic faces using deep learning [first accomplished in 2014 with generative adversarial networks], I said, “Oh, we can use that to generate cells instead.” That’s my job: to model cells. I said, “What if we were able to generate images of cells that came from a certain labeling experiment, and biologists couldn’t tell if those images were real or not?” If we could do that, we would, in some sense, have built a model that understands what that experiment is doing.
Doesn’t that run the risk of seeing things that aren’t really there?
What we’re really trying to do is predict the outcomes of experiments, so scientists can prioritize the experiments that they think are interesting.
Suppose I have an image of a cell, and [the software is] predicting a localization pattern of a thing inside the cell — for example, mitochondria. When we observe mitochondria in our label-free model, what we’re showing is the expected outcome of mitochondrial localization. It’s like the average place where we think those mitochondria are.
Another way to think about it is, say I wanted to run an actual experiment that involved labeling these cells with fluorescent proteins. But instead of running that experiment, all I have are these really, really cheap brightfield microscope images. So I ask the machine to predict the outcome of this labeling experiment. Then, if I see something interesting in the generated image, I can go run that actual experiment.
So are you using AI to help focus experimentation, or replace it?
I think both answers are correct. A scientist says, “The point of an experiment is to prove that your model is wrong.” Because our [deep learning] model is trained totally on data from experiments with fluorescence imaging, that means any time you go out and gather new experimental data that shows me how that model’s wrong, I can add that data to my model to make sure that it’s better next time.

“What we want to be able to do,” Johnson said, “is take a movie of the cell and watch how the relationships between the inner structures change in a predictive sense.”
Chona Kasinger for Quanta Magazine
It’s a win-win situation because either the model is correctly predicting the outcome of experiments, or the model’s getting new data that can give us a better prediction next time.
If you take this process to an extreme, you end up with a machine learning model where you can type into it the parameters of whatever experiment you’d like to run. Then it spits out whatever you’re trying to measure. If you actually go run that experiment, and the real data and the model’s data are the same, then you have a model that understands, in some fundamental sense, what biology is.
Is this approach controversial?
About two or three years ago, people would look at it and say, “I don’t believe you.” I’ve been in conferences where I presented my work and heard something to the effect of, “Get this garbage out of here.” Now people are much more comfortable with this idea. It’s becoming very, very quickly adopted, all over the cell biology imaging universe.
What changed?
My Ph.D. was largely around using classical statistical modeling to do this sort of thing. That’s a very, very powerful tool. But these statistical tools may or may not be able to generate images of cells that are qualitatively realistic. I could make a blurry distribution inside the cell and say, “A brighter spot here is where I would expect to find the mitochondria,” and people would say, “Well, that doesn’t look like a cell at all.” It was very frustrating for me because the math and probability were all correct.
But when we looked at the first images [from the label-free determination model], they looked real. We’re actually seeing where these parts of the cell are. People’s jaws dropped. Then we just ran with the idea.
Seeing is believing?
Yeah, exactly. And the fact that we’re doing it using brightfield imagery just blew everyone away, because in the imaging world, brightfield imagery is largely throwaway data. When we take these images of tissue — just shining normal light on it — that’s just to figure out whether or not the sample is in focus on the microscope. Then it just goes somewhere on a hard drive and nobody ever looks at it again. Experiments using fluorescent molecules are expensive, but brightfield imagery is essentially free. Being able to train [deep learning] models with that expensive data and then using those models to predict where things are inside the cell for the [brightfield] imagery that you took saves a lot of time and money.
You have to train separate deep learning models to recognize the different parts of the cell you’re interested in. Do some work better than others?
Membrane-bound organelles, like the nucleus and mitochondria, are very easy to predict. Other organelles that are not membrane bound, like microtubules or the Golgi apparatus, are very difficult to predict. That almost entirely has to do with the fact that the density of these objects is not very different from that of the surrounding area inside the cell.
How do you get around those limitations?
Instead of using just normal transmitted light, you can use polarized light or other types of imaging that use optical tricks to get different levels of contrast [in the image].
Or, if I’m running an experiment and I can only use three fluorescent labels, I can forget about using them on the structures that the system is already good at predicting and use them instead on ones that are difficult to predict, like actin and microtubules — cytoskeletal structures inside the cell.
You and other scientists at the Allen Institute can continuously improve these models, and the “integrated cell” is based on this work. But can scientists outside the Allen Institute use this method, too?
That’s a big part of our entire mission. When Google built AlphaGo to beat the best Go players in the world, that thing had 200 years of gameplay under its belt. Those resources are totally unattainable by any other institute in the world, except maybe Amazon or Microsoft. We want other people to be able to use our cell lines and our technology for their own research in their labs, but they don’t necessarily need to have that level of refined pipeline that we do.
One of the things we tried very hard to do was to build these models on commodity hardware — just a normal computer with a graphics card — with a number of [training] images that is acquirable by a normal human being in a normal laboratory. All of our models are trained on about 30 images [of fluorescently labeled cell structures]. This is what a graduate student can do in less than an afternoon in their lab. And you can go build a computer that can do this for about $2,000, which is pretty cheap as far as laboratory equipment goes. If you really need to go build a model that does something useful, it shouldn’t be difficult to do.
How do you see this technology progressing? What do you want cell biologists to be able to see with AI’s help in the future?
What we want to be able to do is take a movie of the cell and watch how the relationships between the inner structures change in a predictive sense.
The microtubule and the DNA, for example. When a cell splits in two, the microtubules, which normally make the cell keep its shape, take the DNA and pull it apart into the two copies on either side of the cell. This phenomenon is well known; it’s one of the first things that cell biologists ever saw. But there are a lot of relationships between these two structures that are incredibly subtle — maybe so difficult to detect that a human eye could not see them. We want to be able to use these bleeding-edge computer vision and machine learning methods to automatically dissect the relationships between these structures.
Is this only for image data?
No, we don’t need to limit ourselves. We can take all sorts of signals and measurements of a cell and model their relationships to each other. To use the glass-car metaphor again: You’d not only see all the parts inside distinctly labeled, you’d also see what the mileage is, when it was assembled, how old those parts are, whether or not they’ve been replaced — that sort of thing.
You could think of this [technology] as a data-driven, heads-up virtual reality display for a microscope. Anything that we can measure in our cells, or any pairs of measurements, we can relate to each other. Every five or 10 years, scientists come up with a new way of taking measurements. And it totally changes the way we think about biology, or maybe science in general. When scientists are looking at their tissue sample, we want them to pull up this display and have it predict every single thing about the cells that we could possibly measure.