In the Nucleus, Genes’ Activity Might Depend on Their Location
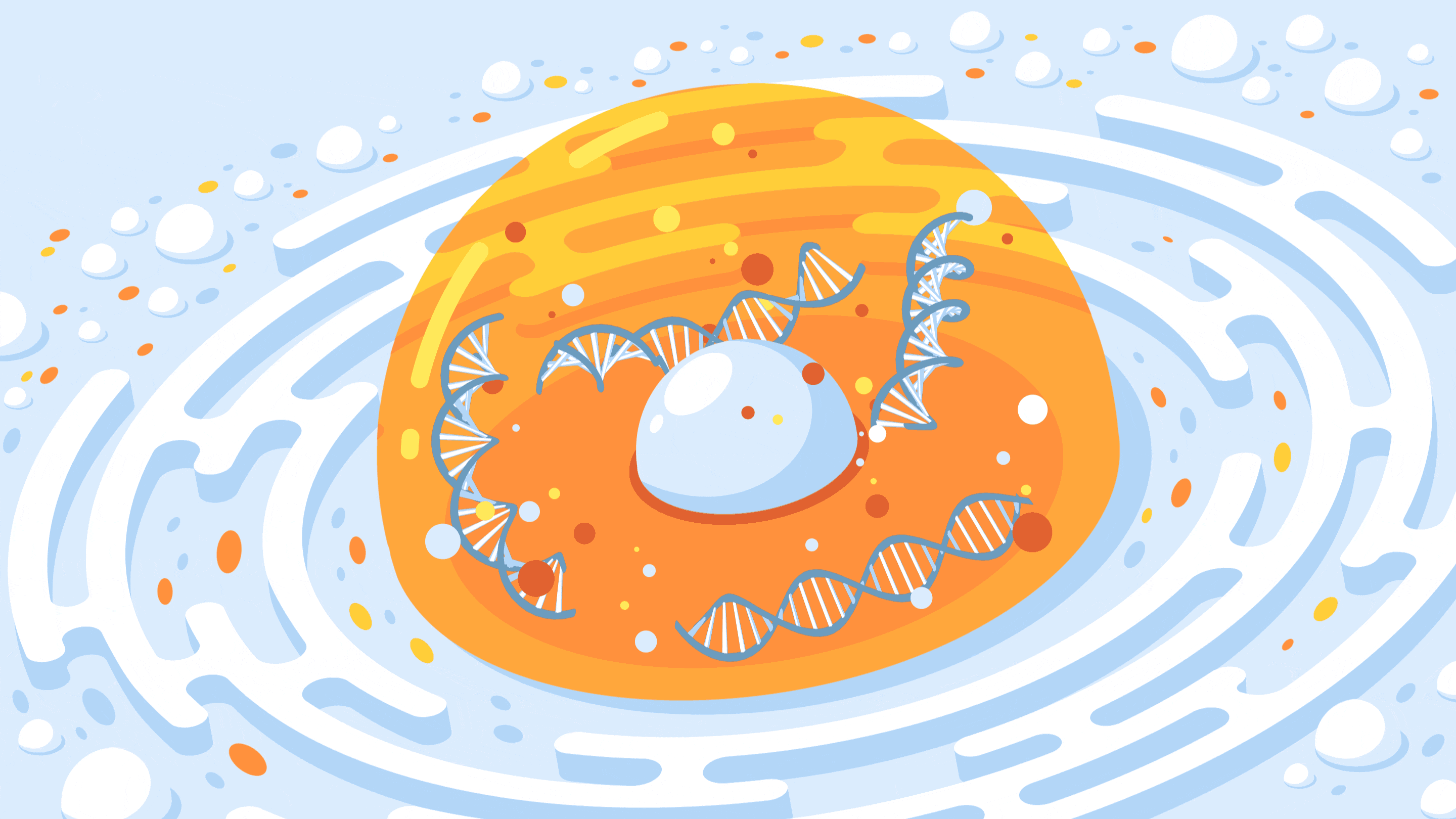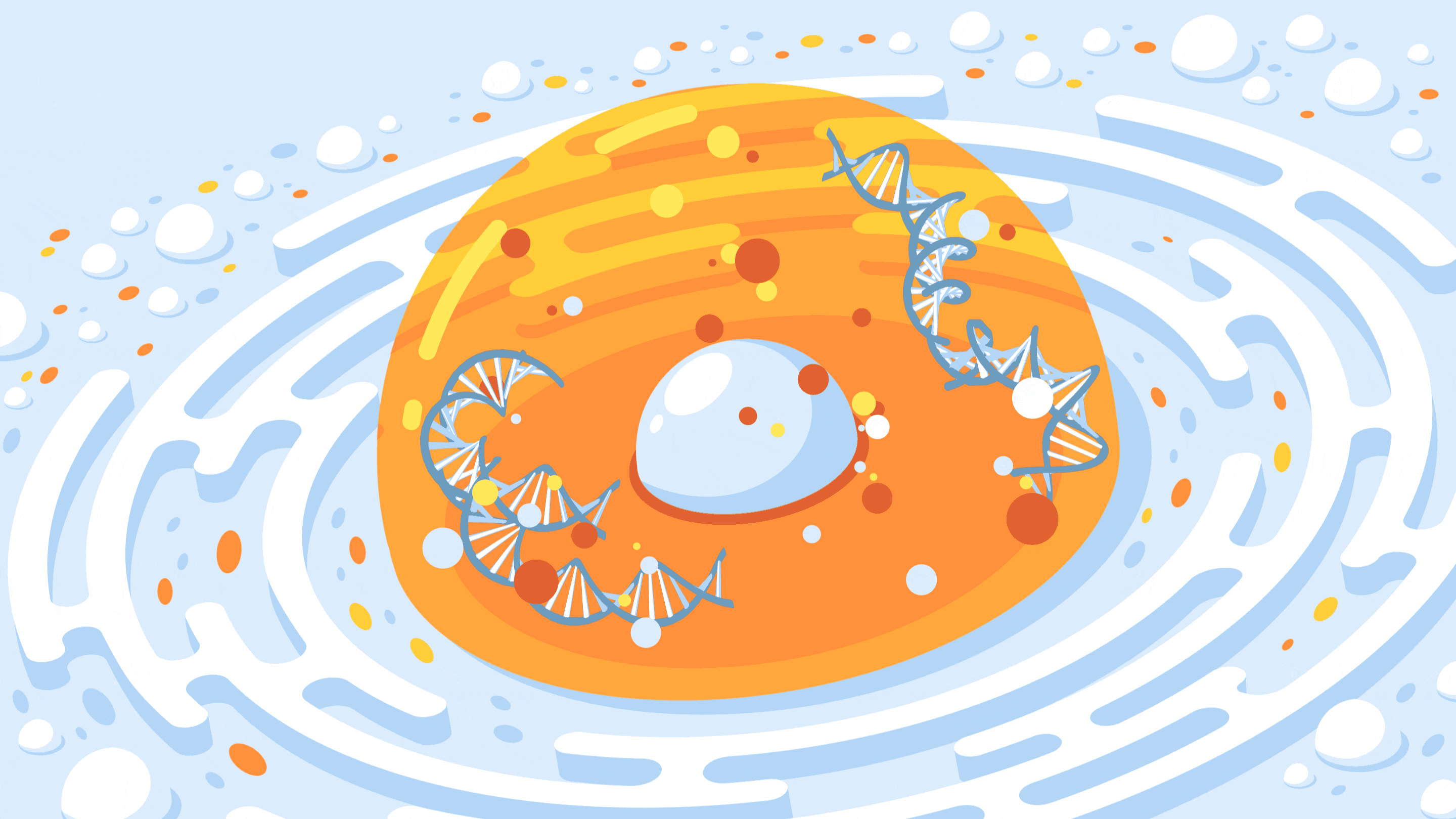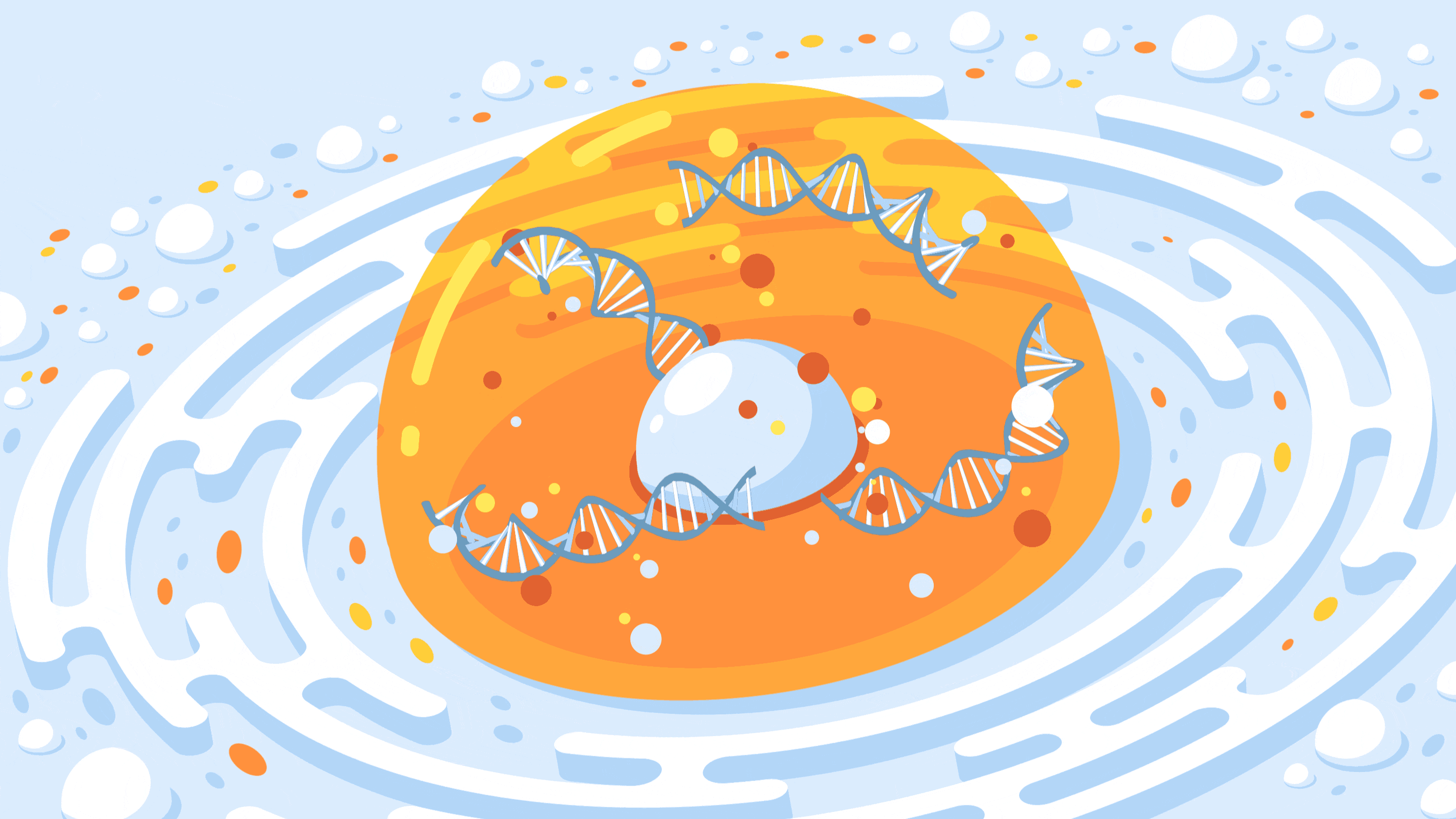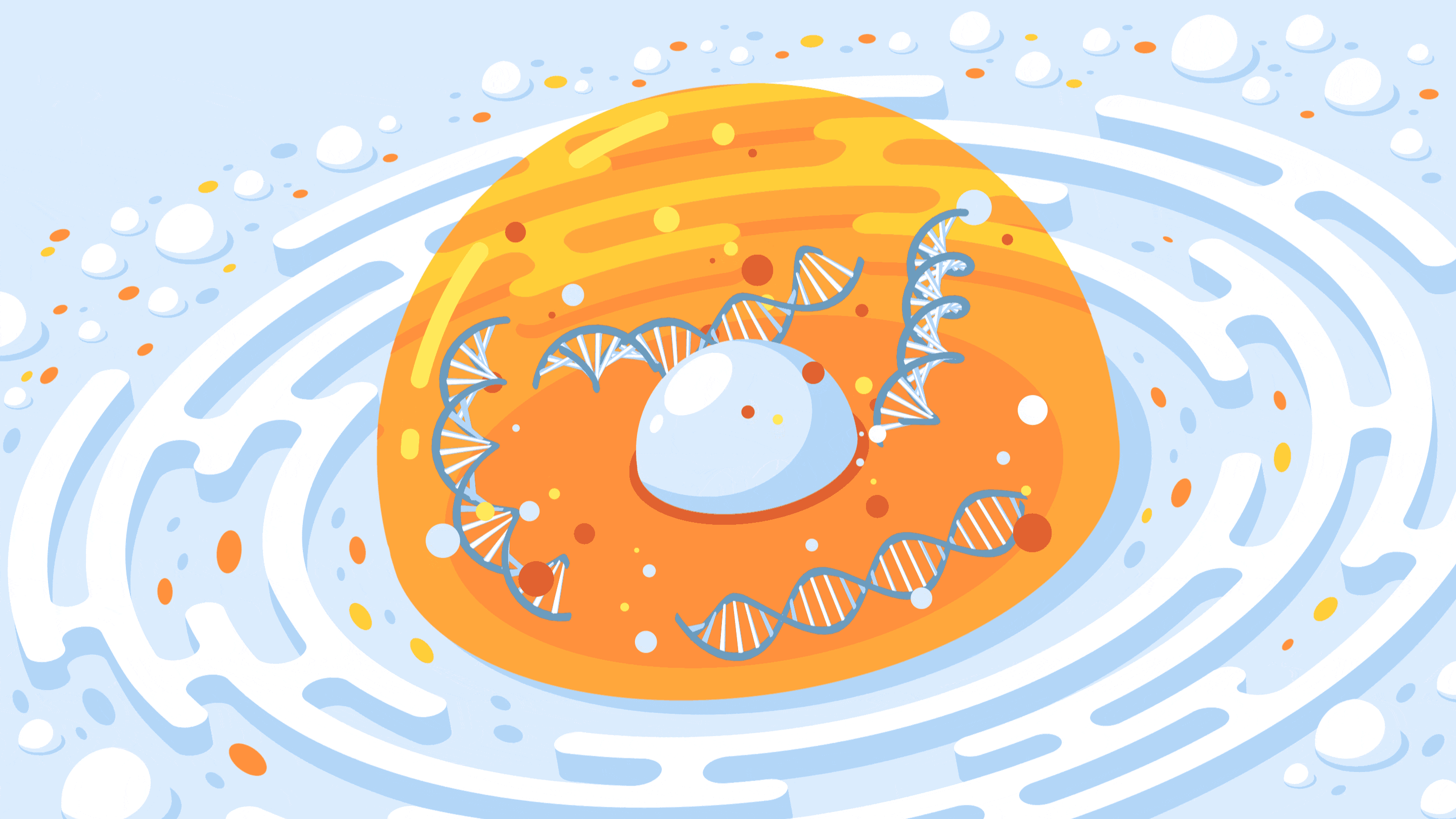
A new CRISPR-based technology allows researchers to control the spatial organization of DNA inside the nucleus of a cell.
Crispe for Quanta Magazine
Introduction
The nucleus of a cell has something in common with a cardboard box full of kittens: People get so fascinated by the contents that they overlook the container. The nucleus itself is often treated as no more than a featureless membranous bag for holding the vitally dynamic genetic material. Yet in fact it has specialized parts and an internal architecture of its own, and scientists have long speculated that precisely how the DNA positions itself with respect to those parts might matter a great deal.
Now a team of researchers is finding credible evidence that this is true and possibly an important influence on gene expression. Using a new technique based on the genome-editing tool CRISPR, they artificially pinned parts of a cell’s DNA to different regions in the nucleus and observed what happened. The work, published last month in Cell, has begun to yield intriguing insights into how various nuclear neighborhoods may relate to gene expression, as either cause or facilitator.
The 6 feet of DNA intricately bundled within a human cell’s tiny nucleus can look as chaotic as a ball of spaghetti or a tangle of thread. But how that DNA gets situated in three-dimensional space is critical — and not at all random. The degree of packing and folding enables genes to be accessible in the right place at the right time, so that the cell’s machinery can find and decode them, dial their activity up or down, and keep everything working as it should. Those rearrangements also put specific parts of the genome near or far from landmarks within the nucleus.
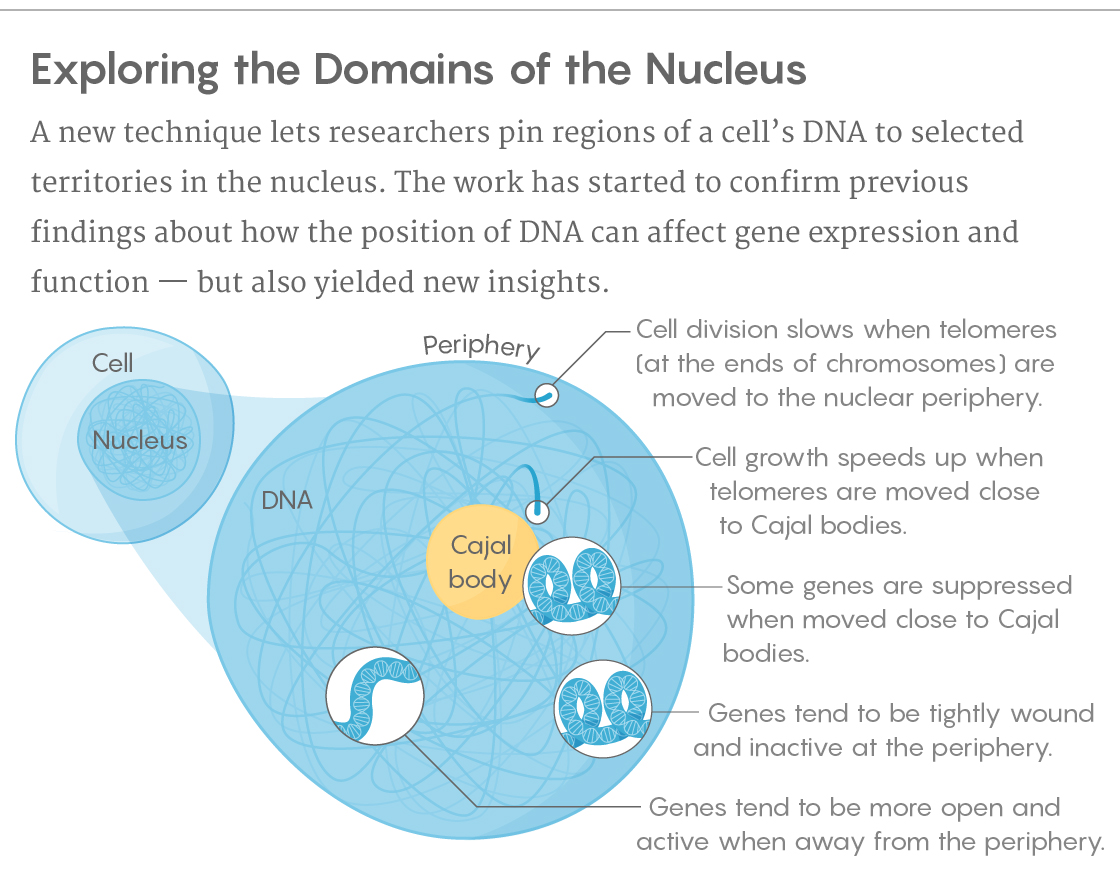
There’s been tantalizing evidence that the positioning of DNA at those nuclear locations may not be coincidental. Tightly wound, silent genes tend to be located toward the periphery of the nucleus, while open, active DNA makes its home toward the interior. During development, as cells differentiate, the DNA reorganizes itself: As some genes shift from a repressed state to an active one, they’ve also been found to move away from the periphery. That said, some other gene regions usually found near the periphery aren’t there all the time, and when they do move, they still show the same levels of activity.
Stanley Qi, a bioengineer at Stanford University, holds a 3D printed model of the modified CRISPR complex that he and his team created to help move DNA to chosen regions of the nucleus.
Biologists have therefore debated how DNA’s condensed structure and expression relate to its nuclear location, and what might be cause rather than effect. Inactive genes with a certain profile might get drawn to the periphery, or the periphery itself may be responsible for silencing them. Those considerations get even more complicated toward the center of the nucleus, which comprises many different domains defined by a variety of nuclear bodies, such as the nucleolus (which assembles ribosomes for protein production) and Cajal bodies (which help to splice RNA). Their functions, too, have been difficult to tease apart: Once again, correlations abound, but pinning down causality is a different story.
“These have been the questions at the epicenter of the studies on the relationship of genome organization and nuclear structure and gene regulation for decades,” said Mitchell Guttman, a biologist at the California Institute of Technology.
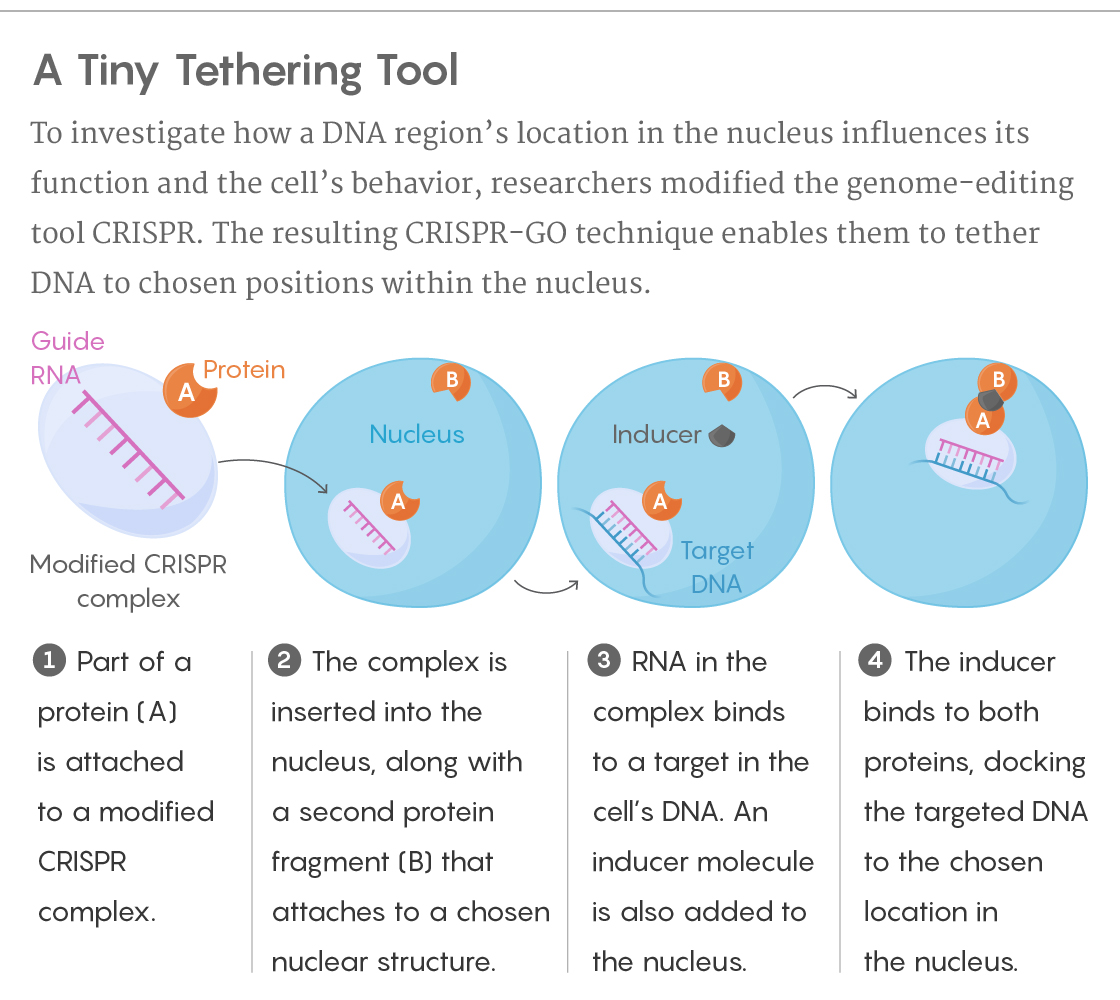
And so for the past four years, Stanley Qi, a bioengineer at Stanford University, and his colleagues have been working on paving a way for scientists to start answering those questions. They turned to CRISPR, a system that has been used widely to edit genes, regulate transcription and take images of cellular processes. Now they’ve innovated a way to harness it for spatial control over the genome. They’ve dubbed the process CRISPR-GO (the GO stands for “genome organization”). “It’s a broad expansion of the CRISPR technology,” Qi explained, “which started five years ago and is still not slowing down.”
The method works a bit like placing a strip of Velcro on the genes the researchers want to move, and another strip on the nuclear body they want to move the genes to. As the DNA passes by, the two corresponding pieces of Velcro end up sticking together.
More specifically, the scientists employ the molecular complex CRISPR/Cas9 — its ability to cut the genome deactivated — to target a specific DNA sequence. Part of a protein is attached to the complex, and a second part is attached to a chosen structure within the nucleus. As the DNA goes about its business, moving and shifting as usual, the researchers add an “inducer” molecule that links to both protein fragments, binding them and tethering the DNA to the new location. Removing the inducer frees the DNA to move away again.
Lucy Reading-Ikkanda/Quanta Magazine
Previously, researchers had used another technique to try to achieve this result. However, they had to engineer a special cell line to integrate a long, highly repetitive bacterial or synthetic sequence next to a gene of interest. The system was difficult to use and limited in scope, and the invasive nature of the method meant the experiment itself could be affecting the results. When such studies yielded inconsistent findings — some concluded that recruiting DNA to the nuclear periphery led to gene silencing, for instance, while others saw no such effect — researchers struggled to interpret them.
But CRISPR-GO doesn’t require scientists to modify the genome, and it can precisely target any region of DNA. “It’s in a way a much gentler approach,” said David Spector, a molecular biologist at Cold Spring Harbor Laboratory in New York, who was not involved in this work. “And more elegant.”
With CRISPR-GO, scientists now have the toolbox to explore what nuclear bodies are really doing: how they may or may not be influencing the activity of specific genes, for one, and what roles they might be playing in health and disease.
Qi and his team showed how that can work. They first noted that different nuclear bodies exhibited their own dynamics. When they repositioned DNA to the nuclear periphery, for example, it was the DNA that did the moving, and the process took nearly a day. But when they relocated sections of the genome to a Cajal body, the latter went to the former within only a few minutes. According to Qi, those findings could reveal information about how long-lasting or transient activation and repression may be in certain parts of the nucleus, and about how actively DNA interactions are maintained.
As previous work had also shown, Qi and his colleagues observed that a spatial relationship seemed to exist for some genes and not others. Still, they uncovered some surprising links, including long-range effects. Moving some protein-coding genes to a Cajal body, for example, suppressed their activity — as well as that of DNA located hundreds of kilobases away.
The team also looked at noncoding regions of genes, sequences with regulatory or other functions that make up the vast majority of DNA. They focused specifically on telomeres, sections of DNA located at the tips of chromosomes that relate to cellular life span. When the telomeres were repositioned to the nuclear periphery, cell division ground almost to a halt. Moving the telomeres close to the Cajal bodies, though, had just the opposite effect, causing cells to grow and divide much more rapidly.
Lucy Reading-Ikkanda/Quanta Magazine
“This implies that the location of the telomeres in the nucleus is important for cells to finish their proper cell cycle,” Qi said. He speculates that the Cajal bodies may have had this effect because they’ve previously been shown to produce an enzyme that helps maintain the length of telomeres. “We think we were potentially co-localizing the manufacturing plant with the consumer market,” he said.
But the researchers still need to root out just why these effects occurred. They’ll have to perform further experiments — targeting various genes and nuclear bodies in diverse cell types, and testing not only for effects on gene expression but also on genomic stability and other factors — to find out why and how the genome is organized as it is. At the very least, it seems to “build in an extra level of control,” Guttman said. “By creating active and inactive territories, the nucleus can prevent proteins that silence transcription from aberrantly turning off a gene that needs to be on, and vice versa.”
Susan Gasser, a molecular biologist at the Friedrich Miescher Institute for Biomedical Research in Switzerland, thinks experts will find that location in the nucleus is important for very particular processes, such as DNA repair — but that a lot of the time, “it instead fine-tunes gene expression.” The open or condensed state of the DNA itself may be more influential. Still, CRISPR-GO can be used to test that idea, she said.
It can also help investigate the role of nuclear organization in development and disease. Pathologists have been using nuclear morphology as a diagnostic tool for a long time: Altered states and distributions of DNA correlate with cancer and other conditions, as does an increase in the number of certain nuclear bodies. But it’s been unclear if those are the results of the disease or its cause.
Now Qi feels that he and others are in a position to find out. One day, he said, the system might be used not just for exploration and basic research, but as a means of treatment as well.
Even so, some experts still have reservations. Andrew Belmont, a cell biologist at the University of Illinois at Urbana-Champaign, cautions that the researchers still need to confirm that their technique accurately reflects natural processes in the cell, and not some artificial consequence of the tethering procedure. He and his colleagues, along with a few other groups, have developed alternative systems to get around that concern, which involve inserting natural sequences into targeted DNA regions that are already ordinarily associated with one nuclear body or another. Still, he agrees that CRISPR-GO represents a major step forward.
Guttman concurred. “I anticipate that this will become an incredibly powerful tool for many in the field to start deciphering those really old and really important questions,” he said.





