Researchers Tap a Sleep Switch in the Brain

Two genes found in mice influence sleep and dreaming.
Jessica Florence
Introduction
For more than a century, biologists have been studying sleep. And over the decades, interesting findings have piled up: Sleep deprivation, we now know, can be lethal. Sleep pressure — the need for sleep — increases the longer you’re awake. In humans and other mammals, sleep is very recognizable in readings of brain activity. Early on, the squiggling line of an EEG readout starts to form kinks called sleep spindles, unfurling across the glowing screens inside sleep labs.
But for all these meticulous observations, the molecular machinery behind our nightly departure from consciousness remains one of the deepest mysteries in modern biology. “On a mechanistic level,” said Amita Sehgal, a chronobiologist at the University of Pennsylvania who has pioneered the study of sleep using fruit flies, “we don’t know what’s happening to make us sleepy.” Biologists have mutated thousands of lab organisms like flies, picked out those with sleep abnormalities, and studied their genes, as well as those of people with sleep disorders. The idea is that if we can identify which genes are disrupted — what is making people fall asleep at their desks, or what makes flies too snoozy to mate — we’ll be closer to understanding why we get sleepy and what, on the molecular level, sleep consists of.
Now, a team of researchers in Japan and Texas have unveiled the first results of a colossal experiment that takes this approach with mice, on a scale never seen before: Over the course of several years, the team mutated the genes of thousands of mice, hooked them up to brain wave monitors and watched them sleep. In a recent paper in Nature, the researchers revealed two genes that seem to be involved in sleep’s biological machinery. The first was found in mice that need much more sleep than normal. The other comes from mice that spend less time in dreaming, or REM, sleep. The results suggest new leads for sleep researchers to chase, and they contribute to a rising tide of research dissecting the molecular underpinnings of sleep.
The idea for the experiment came in 2007 or so, said Masashi Yanagisawa, a biologist who at the time was at the University of Texas Southwestern Medical Center. He had stumbled into the sleep field about a decade before, when he and his colleagues, on a hunt for interesting neurotransmitters, identified a protein that turned out to be involved with narcolepsy. Sufferers of the condition can go from chatting and laughing to REM sleeping in seconds. Studies with mice lacking the protein and a colony of narcoleptic dogs at Stanford, as well as human research, soon established the protein’s role. Called orexin, it helps keep people awake — but that was far from the whole story, as researchers uncovering the circuits of neurons behind alertness and sedation knew. Yanagisawa grew more and more interested in cracking open the black box of sleep.
Biologists had recently begun screening thousands of mutant flies in search of sleep genes. In flies, which are in constant motion when they’re awake, researchers define “sleep” as periods of inactivity. But the gold standard for studying human sleep is electroencephalography, or EEG, which lets observers track the stages of sleep by measuring brain activity. It would be interesting to try it with mice, Yanagisawa thought, which, while wildly more expensive than flies — they take much longer to breed and take up far more space and resources — can be fitted with implants to allow EEG. And mice are evolutionarily closer to humans as well. He conferred with Joseph Takahashi, a colleague at UT Southwestern who specialized in immense mouse studies.
Each week, they decided, they could mutate maybe 30 or 40 mice using a drug, implant brain-activity monitors in their heads, and see how long they slept and how long they spent in each phase of sleep. The study generated data like those from human sleep studies. Mice with abnormalities could be picked out from their sleeping brain activity, and their genomes sifted through to find the genes responsible.
The pilot phase began with just a few hundred animals. “Each mouse could be a treasure,” Yanagisawa said. “You just have to find out which one.” Early on they were encouraged by the discovery of one mouse with sleep abnormalities. And in 2009, Yanagisawa received an enormous grant from the Japanese government to study sleep. The grant was so large that it enabled him to set up a facility at Tsukuba University in Japan to continue the screening on a grand scale. “Now we do up to 100 mice a week,” Yanagisawa said.
Lucy Reading-Ikkanda/Quanta Magazine
The Nature paper reports the results from the first 8,000 mice. For years now, Sehgal and many other sleep researchers say, they’ve been hearing updates on the team’s progress at meetings. Yet the identity of the genes has always been a closely kept secret. Sehgal recounted a meeting at which, during an informal presentation session, a researcher from the Yanagisawa lab teasingly told the audience, “Ask me anything you want!”; the crowd, a few drinks in to the evening, roared back, “What are the genes?”
“So I was excited to see the paper,” Sehgal said with a laugh, “and finally find out what these genes were.”
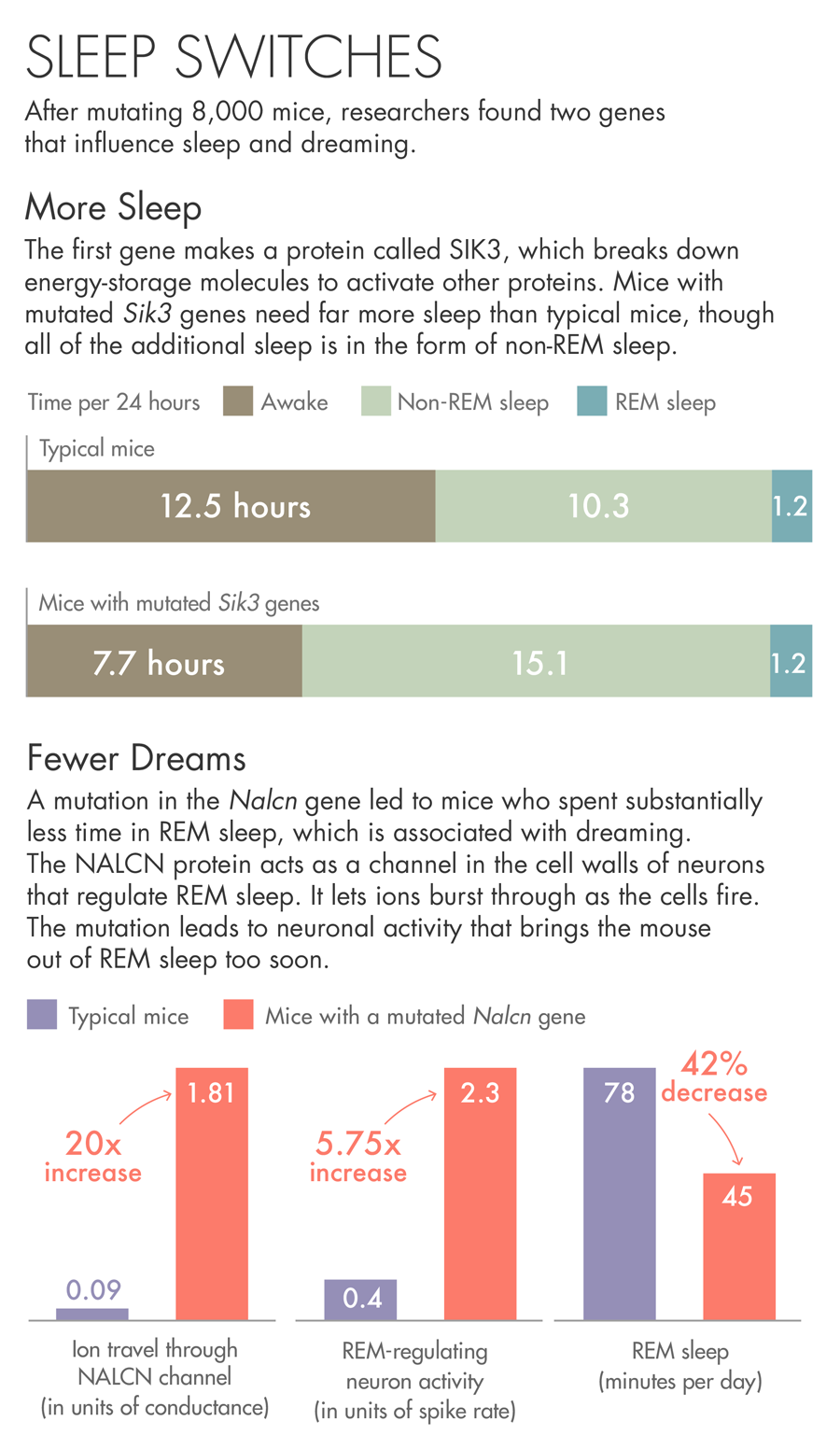
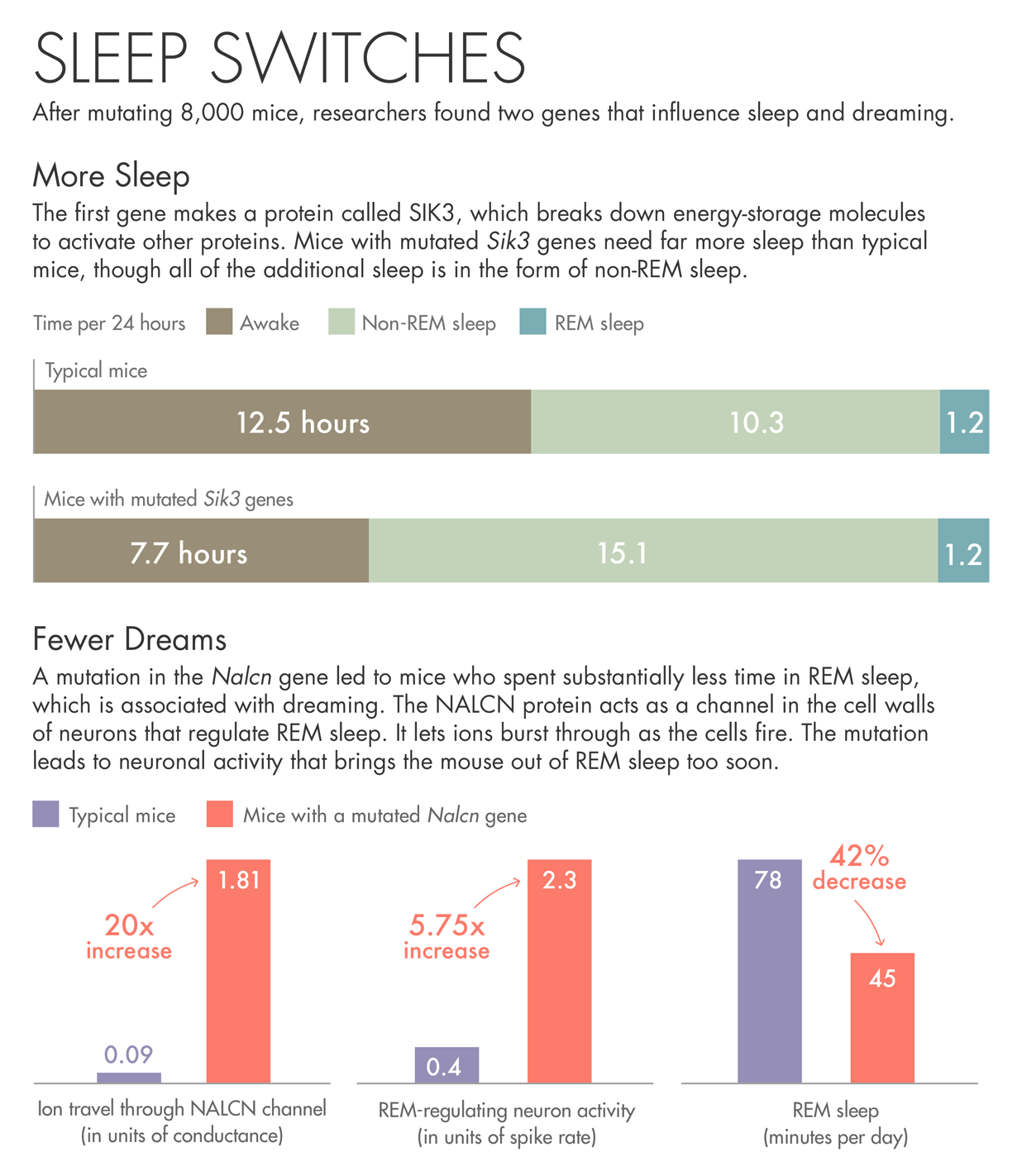
The first gene the team describes makes a protein called SIK3. Mice with a mutated Sik3 gene spend little time awake — each day they spent more than 30 percent more time asleep than other mice. SIK3 is an enzyme that uses up the cell’s batteries — energy-storage molecules called ATP — to activate other proteins. When the researchers investigated the nature of the mutation, they found that it had removed SIK3’s on-off switch. They believe the mutation jams SIK3 in the on mode.
The other gene came from a mouse that spent less than the usual amount of time in REM sleep, bobbing randomly in and out of a state that usually lasts for a predictable period of time. The mutation in this case was in a protein called NALCN, which acts as a channel in the cell walls of neurons. The protein lets ions burst through as the cells fire. The mutation in this particular mouse may be causing neuronal activity that brings the animal out of REM sleep too soon.
Identifying genes related to sleep doesn’t lead to answers as quickly as one might hope, however. Sleep is so crucial to survival that the ancestors of every sleeping creature alive today risked death to do it daily. That means that there are likely many ways to ensure that it happens, a redundancy that makes it difficult to dissect in experiments. A mutant fly strain that starts out with a severe sleep abnormality will evolve over the generations to sleep more and more normally as time goes on, Sehgal said. That can be frustrating for researchers, though it provides evidence of sleep’s robustness. And most of the genes linked with sleep in the last decade or so of studies likely have many purposes. “There is not something called a ‘sleep gene,’” said Divya Sitaraman, a sleep researcher at the University of San Diego. “There are these genes that have very basic functions that also affect sleep.”
This new paper provides a great example. SIK3, as it happens, doesn’t show up in just the brain. Over the years researchers have found it being used all over the body. It has been linked to metabolic problems, which can make organisms sleepy, said Tom Scammell, a doctor and sleep researcher at Harvard Medical School. A medical work-up of the mouse might help determine whether its drowsiness is because SIK3 affects sleep directly or because of damage to the liver or kidneys, he said.
Still, as work in animal models has continued to churn out these sleep-related genes, the collection of genes linked to sleep in humans has been kicked up a notch. Some researchers have used vast sets of data — thousands and thousands of people, sorted according to how much time they spend sleeping or whether they get up early or late — to see whether these can be linked to specific genetic variations. At least three such studies have been published in the last year.
So far, not many of the associations of genes to sleep have been found by more than one study, said Richa Saxena, a geneticist at Harvard Medical School, Massachusetts General Hospital and the Broad Institute who is an author of one of those studies. That’s likely the redundancy of sleep raising its head again — almost no genes are so strongly connected to sleep that their influence can rise above the noise. But she is hopeful that these statistical associations, paired with the results of screening studies, can bring knowledge of the mechanisms of waking and sleep into reach. For instance, the NALCN channel complex includes a protein that Saxena’s work has linked with sleep.
Finding those connections will make all the difference, said Dragana Rogulja, a fruit fly sleep researcher at Harvard Medical School. She was pleased to learn recently that a sleep-related protein she’d discovered showed up in the same cells as another sleep-related protein identified by another lab. Even though the significance of the link isn’t clear, it’s a step forward. “I was pretty happy about that,” she said, “because most of this stuff is not really connected yet.
“But this is normal in a field that is relatively new,” she said. Yes, sleep research is an old field, she said, “but it was pretty descriptive. I think it’s a really exciting time in sleep research right now, because we’re able to move from that very descriptive, poetic style to a mechanistic one.”
That transformation will continue as researchers delve more deeply into the private life of each protein uncovered by genetic screens and by human studies. Yanagisawa and collaborators are in the thick of characterizing both proteins — SIK3 and NALCN — working on the questions of where they are active and what they interact with. The group continues to mutate mice and watch them sleep; they expect to publish further findings soon. “I feel like these last several years have been the funnest part of my career,” Yanagisawa said. “It’s really exciting.”





