Simple Gene Circuits Hint at How Stem Cells Find New Identities
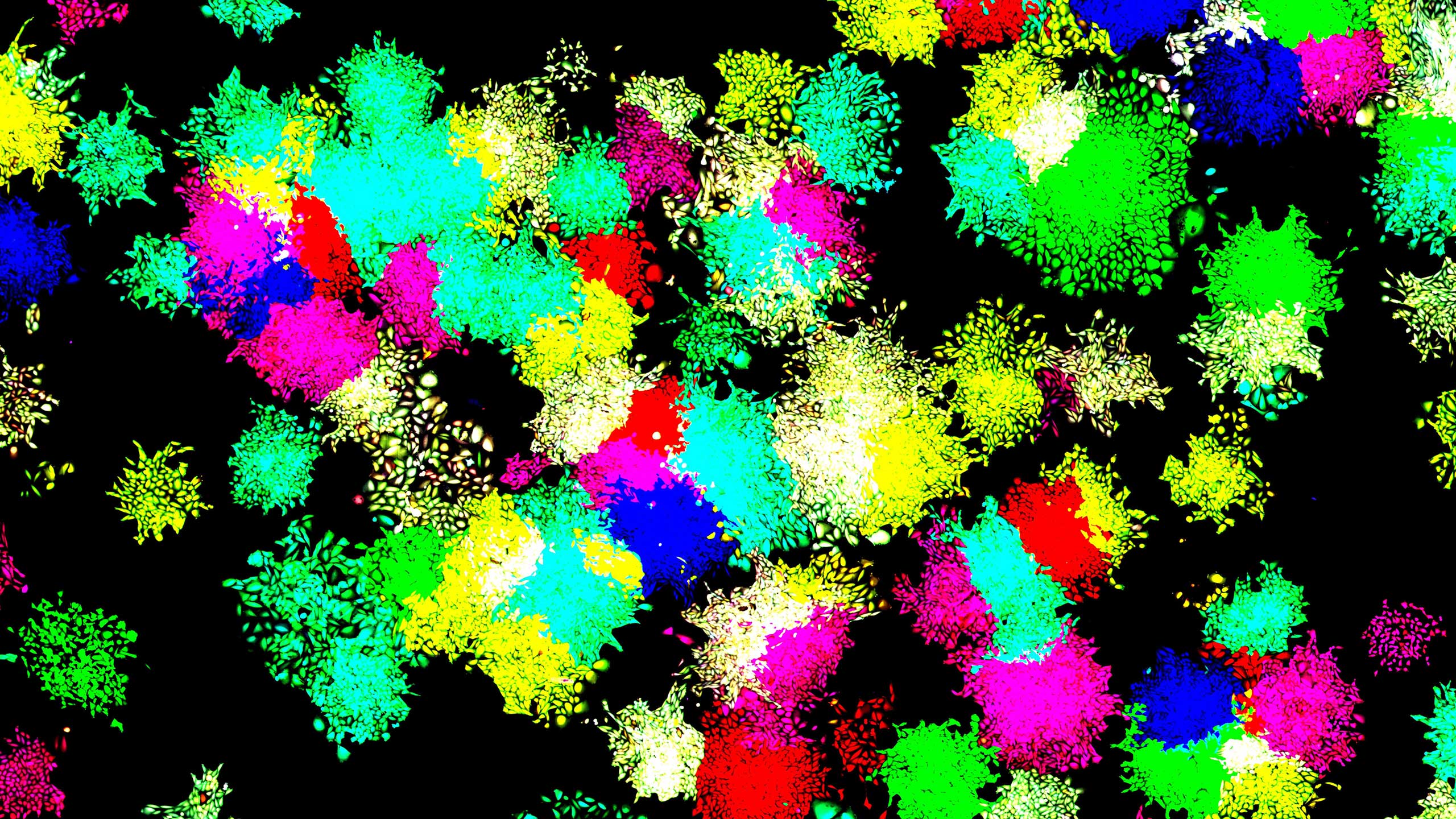
All the cells in this culture are genetically identical, but individual colonies glow in different colors because a synthetic system of regulation keeps them in distinct states.
Ronghui (Ron) Zhu / Elowitz lab / Caltech
Introduction
The human body contains more than 200 types of cells by most estimates, all descended from a single fertilized egg. The spindly cells of the skin, the gangly branching neurons, the plump fat cells, the exquisitely sensitive rods and cones of the eye — all of these are products of a long process of development, during which their physical forms altered beyond recognition. With few exceptions, however, all these cells carry the same genes as that fertilized egg. The only thing that changes from cell to cell is which genes are active.
But how do genetically identical cells get shunted into different identities? What happens at the molecular level to turn stem cells into skin cells, and why do they stay that way instead of morphing into muscle or fat?
Researchers have struggled to answer such questions, which are relevant to the development of all complex organisms, whether they are mustard plants, centipedes or blue whales. Early attempts at genetic models always lacked important aspects of what biologists saw in nature — not least a simplicity that would allow them to scale up to define multitudes of cell fates.
Now, a group of biologists at the California Institute of Technology with backgrounds in physics have reported in Science that they have devised a simple network of genes that gives rise to surprisingly complex, lifelike behaviors. It could represent an important advance in understanding how nature tells cells to differentiate.
By introducing a small number of engineered genes into cells and applying the right chemical cues, the researchers were able to direct the cells into seven different stable states, each distinguishable under the microscope by a different glowing color. The cells exhibited key properties associated with differentiated cells; for example, they were stably committed to being one type of cell, but they also exhibited a “memory” of their previous activity that affected their responses to new circumstances.
Mathematical models suggest that with just a few more genes, it might be possible to define hundreds of cellular identities, more than enough to populate the tissues of complex organisms. It’s a finding that opens the door to experiments that could bring us closer to understanding how, eons ago, the system that builds us was built.
The Limits of Mutual Repression
Developmental biologists have illuminated many tipping points and chemical signals that prompt cells to follow one developmental pathway or another by studying natural cells. But researchers in the field of synthetic biology often take another approach, explained Michael Elowitz, a professor of biology and bioengineering at Caltech and an author of the new paper: They build a system of cell-fate control from scratch to see what it can tell us about what such systems require.
More than 20 years ago, researchers at Boston University led by James Collins took a major step in that direction. In a Nature paper, they described an artificial cell control circuit they had inserted into Escherichia coli bacteria. The circuit could flip the cells between two states. The engineered cells glowed green as long as a gene that repressed production of the fluorescent pigment was turned off. However, if the scientists added one chemical to the cell’s culture solution, the repressor gene was activated and the color disappeared. A dose of another chemical reversed the process by forcing a second repressor into action, bringing the green back.
These two states were stable: Until a chemical was added to trigger a change, a cell remained either glowing or dark. This stability recalled the behavior of cells in nature whose fates are permanently set by chemical commands issued during development. The key to the control system was that the two repressors repressed each other — when one was ascendant, the other was dormant.
This kind of mutual repression has been central to most cell control systems that biologists have devised since Collins’ experiment. An elegant example is a system designed by Xiao Wang, an associate professor of biomedical engineering at Arizona State University, and his colleagues, presented in a 2017 paper. Using two mutually repressing transcription factors and two genes that activated themselves, they created a system capable of putting E. coli into four different states.
Mutual repression, however, can be complex to scale up. You can generate additional stable states by adding more genes, but each gene must then inhibit all of the others. Such an approach seemed cumbersome even for ambitious synthetic biology experiments, and nature clearly couldn’t rely on such a brittle system to direct the development of cells in organisms.
Flexible, Adaptable and Robust
When Elowitz discussed this problem with Ronghui Zhu, a graduate student in his lab, they wondered whether there could be a simpler way to scale up. “A fundamental property of the natural system is that it is scalable. That was, I think, a key insight,” said Elowitz. “You need a design that [doesn’t require] that every time you add something, you have to reengineer everything that’s already been put in.”
They looked for a solution in nature, where transcription factor proteins have been seen binding together in pairs, or dimers. A transcription factor will sometimes bind to a copy of itself, and sometimes to a completely different factor. What it’s bound to can radically alter a factor’s abilities, allowing it to activate new genes or turning it off entirely. The permutations of these pairings form a network of possible states for a cell.
So Elowitz and Zhu worked out the math describing a system in which pairs of transcription factors inhibiting and promoting each other could control cell state. In their new system, each gene produces a transcription factor protein; these proteins bind together as dimers to exert an effect. If two copies of the same factor bind together, the resulting “homodimer” stimulates further protein production by the gene — a positive feedback loop. If a factor binds to a different factor to form a “heterodimer,” however, the transcription factor is inactive.

Michael Elowitz and Ronghui Zhu, researchers in biology and bioengineering at the California Institute of Technology, devised a simple, modular system of genes and transcription factors to control mammalian cells in culture.
What’s clever about this arrangement is that once the activity or inactivity of a transcription factor gene is set, it tends to stay that way. The precise amounts of different dimer combinations can be fine-tuned to produce any desired pattern of gene activity. Significant environmental changes that affect the stability of the transcription factors can flip a cell from one state to another, but random small changes are shrugged off. This makes the control system flexible, adaptable and robust.
Much later, Elowitz, Zhu and their team decided to see whether they could actually build it in mammalian cells, a change from other labs’ systems that were mainly built in E. coli. Drawing on a toolbox of transcription factor components assembled by their colleagues, they designed two that were active as homodimers but inactive as heterodimers. The model suggested that a control system built around these could put cells into three distinct states: one in which only the first gene was active, one in which only the second gene was active, and one in which both were active. Which state predominated would depend on the stability of the factor proteins and how likely they were to bind to each other. “The design gives you both combinatorial control and also enables one state to repress the other states,” said Zhu.
Of course, modeling is one thing; making something work in a living cell is another. “If you do a lot of mathematical modeling of biology, you always know that the models are a very rough approximation to what is going on in many cases, and it’s often challenging to make them predictive,” said Elowitz. The team was surprised, then, when Zhu introduced the genes for the initial test into hamster cells and the cells obediently morphed into a painterly pattern of green, red and yellow. The system, which they named MultiFate, seemed to work.
To see if it would scale as the model predicted, they added a third transcription factor, which brought the expected number of states up to seven. The cells obligingly developed a kaleidoscope of seven hues. If they were left undisturbed, the states persisted for over a month, echoing the stability of a natural system.
Merrill Sherman/Quanta Magazine
The researchers also observed how the cells responded to change. By altering the concentration of a chemical in the cells’ environment, they could destabilize the transcription factor proteins; as predicted, this caused the cells to move between states. Intriguingly, though, the responses of the engineered cells were shaped in part by their histories. They switched states when the concentration of the chemical agent went from high to low — but when the concentration was raised again, they didn’t just revert.
This behavioral asymmetry has a parallel in nature, where cells that live through a time of scarcity, for instance, may stay in a permanent state of hoarding energy. Resetting the environment doesn’t wipe away the cell’s experience.
Walking Through the Cellular Landscape
“It’s extremely clever,” said Ahmad Khalil, a professor at Boston University who co-authored a commentary about the Elowitz and Zhu paper for Science. “[It] demonstrates how you can walk through this landscape — reshape this landscape — of different states by just increasing or decreasing protein stability.”
That’s a profound accomplishment, because whatever process led to the seething complexity of enormous multicellular organisms today must have started off quite simple, and it very likely relied on something basic and mutable like protein stability. The system that Elowitz and Zhu described suggests that principles like these would have been sufficient to generate the great variety we see in nature.
Wang, who developed the E. coli system with four stable states, thinks it’s telling that the researchers’ modeling drew on nonlinear dynamics, a branch of mathematics that deals with systems that often have complex, surprising outcomes. “The whole gene regulation network is a nonlinear network,” he said. And while nonlinearity can often lead to chaos, in biology it typically does not. “So there must be something else in there, some deep, deep principles and rules to make the thing so complex but also so robust.”
“Theoretically, if you have a master network of eight transcription factors, you have the core machinery to have all the possibilities to form a human body,” Wang added. Indeed, Elowitz and Zhu wrote in their MultiFate paper that with just 11 transcription factors, it should be possible to produce more than 1,000 steady states.
“On paper it’s very scalable,” Wang said. “But when you scale up, it needs to be cautious. In biology there is so much that’s unknown. We might see something we don’t expect.”
Going forward, Khalil speculated, researchers could hope to use the MultiFate system to control real aspects of a cell’s growth and change, and not just its color. Perhaps cells introduced into patients could be engineered to respond to their environment by following desirable developmental paths. If they sensed cancer, for example, they might develop in a diagnostically or therapeutically useful way. “It’s a very cool notion,” he said.
For Elowitz, the system is a doorway into understanding the teeming weirdness of biology as more than just a Rube Goldberg machine. The artist’s whimsical contraptions, which performed simple tasks with the maximum number of steps, were “the perfect embodiment of the unevolvable design,” he said — of something that does what it’s meant to do but nothing else.
“Natural systems … may look like that superficially because we don’t fully understand what’s going on,” he said. “Once we understand the right way to look at it, we can hopefully appreciate it as a simple design.”



