The Math That Tells Cells What They Are

Cells in embryos need to make their way across a “developmental landscape” to their eventual fate. New findings bear on how they may do this so efficiently.
Adrian du Buisson for Quanta Magazine
Introduction
In 1891, when the German biologist Hans Driesch split two-cell sea urchin embryos in half, he found that each of the separated cells then gave rise to its own complete, albeit smaller, larva. Somehow, the halves “knew” to change their entire developmental program: At that stage, the blueprint for what they would become had apparently not yet been drawn out, at least not in ink.
Since then, scientists have been trying to understand what goes into making this blueprint, and how instructive it is. (Driesch himself, frustrated at his inability to come up with a solution, threw up his hands and left the field entirely.) It’s now known that some form of positional information makes genes variously switch on and off throughout the embryo, giving cells distinct identities based on their location. But the signals carrying that information seem to fluctuate wildly and chaotically — the opposite of what you might expect for an important guiding influence.
“The [embryo] is a noisy environment,” said Robert Brewster, a systems biologist at the University of Massachusetts Medical School. “But somehow it comes together to give you a reproducible, crisp body plan.”
The same precision and reproducibility emerge from a sea of noise again and again in a range of cellular processes. That mounting evidence is leading some biologists to a bold hypothesis: that where information is concerned, cells might often find solutions to life’s challenges that are not just good but optimal — that cells extract as much useful information from their complex surroundings as is theoretically possible. Questions about optimal decoding, according to Aleksandra Walczak, a biophysicist at the École Normale Supérieure in Paris, “are everywhere in biology.”
Biologists haven’t traditionally cast analyses of living systems as optimization problems because the complexity of those systems makes them hard to quantify, and because it can be difficult to discern what would be getting optimized. Moreover, while evolutionary theory suggests that evolving systems can improve over time, nothing guarantees that they should be driven to an optimal level.
Yet when researchers have been able to appropriately determine what cells are doing, many have been surprised to see clear indications of optimization. Hints have turned up in how the brain responds to external stimuli and how microbes respond to chemicals in their environments. Now some of the best evidence has emerged from a new study of fly larva development, reported recently in Cell.
Cells That Understand Statistics
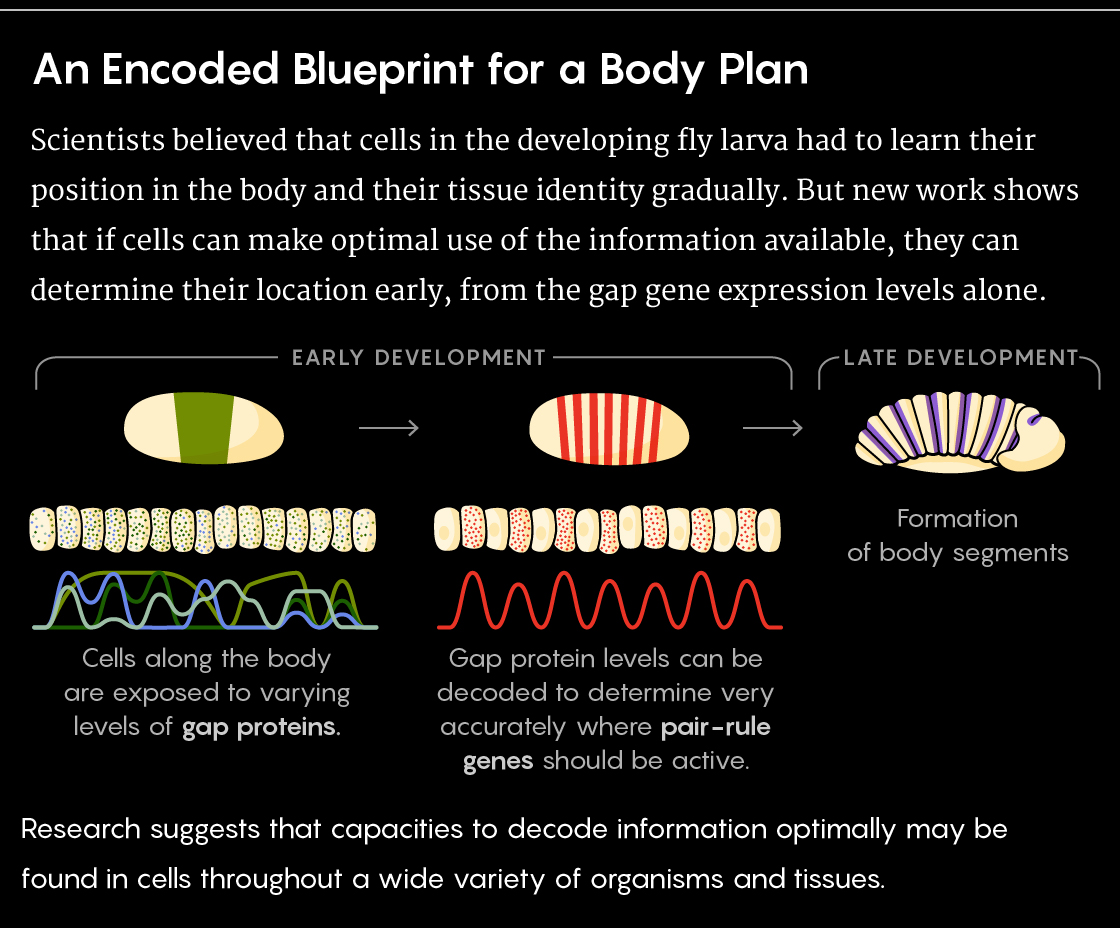
For decades, scientists have been studying fruit fly larvae for clues about how development unfolds. Some details became apparent early on: A cascade of genetic signals establishes a pattern along the larva’s head-to-tail axis. Signaling molecules called morphogens then diffuse through the embryonic tissues, eventually defining the formation of body parts.
Particularly important in the fly are four “gap” genes, which are expressed separately in broad, overlapping domains along the axis. The proteins they make in turn help regulate the expression of “pair-rule” genes, which create an extremely precise, periodic striped pattern along the embryo. The stripes establish the groundwork for the later division of the body into segments.
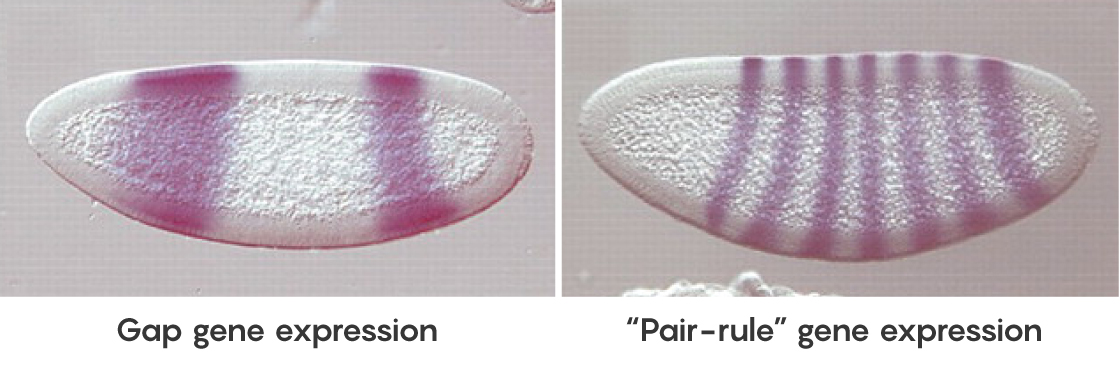
Early in the development of fruit flies, four “gap” genes are expressed at different levels along the long axis of the larval body. That pattern lays the foundation for the expression of “pair-rule” genes in periodic bands later, which give rise to specific body segments. The purple stain in the embryo at left shows the expression of one gap protein; the staining in the later larva at right reveals one pair-rule protein.
How cells make sense of these diffusion gradients has always been a mystery. The widespread assumption was that after being pointed in roughly the right direction (so to speak) by the protein levels, cells would continuously monitor their changing surroundings and make small corrective adjustments as development proceeded, locking in on their planned identity relatively late. That model harks back to the “developmental landscape” proposed by Conrad Waddington in 1956. He likened the process of a cell homing in on its fate to a ball rolling down a series of ever-steepening valleys and forked paths. Cells had to acquire more and more information to refine their positional knowledge over time — as if zeroing in on where and what they were through “the 20 questions game,” according to Jané Kondev, a physicist at Brandeis University.
Such a system could be accident prone, however: Some cells would inevitably take the wrong paths and be unable to get back on track. In contrast, comparisons of fly embryos revealed that the placement of pair-rule stripes was incredibly precise, to within 1 percent of the embryo’s length — that is, to single-cell accuracy.

Thomas Gregor, a biophysicist at Princeton University, has been carefully collecting data on developing fly embryos in his lab for more than 12 years, in the hope of uncovering an underlying physical principle — “a physicist’s dream,” he calls it.
Theodore H. Lewis III
That prompted a group at Princeton University, led by the biophysicists Thomas Gregor and William Bialek, to suspect something else: that the cells could instead get all the information they needed to define the positions of pair-rule stripes from the expression levels of the gap genes alone, even though those are not periodic and therefore not an obvious source for such precise instructions.
And that’s just what they found.
Over the course of 12 years, they measured morphogen and gap-gene protein concentrations, cell by cell, from one embryo to the next, to determine how all four gap genes were most likely to be expressed at every position along the head-to-tail axis. From those probability distributions, they built a “dictionary,” or decoder — an explicit map that could spit out a probabilistic estimate of a cell’s position based on its gap-gene protein concentration levels.
Around five years ago, the researchers — including Mariela Petkova, who started the measurement work as an undergraduate at Princeton (and is currently pursuing a doctorate in biophysics at Harvard University), and Gašper Tkačik, now at the Institute of Science and Technology Austria — determined this mapping by assuming it worked like what’s known as an optimal Bayesian decoder (that is, the decoder used Bayes’ rule for inferring the likelihood of an event from prior conditional probabilities). The Bayesian framework allowed them to flip the “unknowns,” the conditions of probability: Their measurements of gap gene expression, given position, could be used to generate a “best guess” of position, given only gap gene expression.
The team found that the fluctuations of the four gap genes could indeed be used to predict the locations of cells with single-cell precision. No less than maximal information about all four would do, however: When the activity of only two or three gap genes was provided, the decoder’s location predictions were not nearly so accurate. Versions of the decoder that used less of the information from all four gap genes — that, for instance, responded only to whether each gene was on or off — made worse predictions, too.

William Bialek, a Princeton biophysicist, believes that optimization principles constitute “a very concrete idea” that could help experts better understand biological systems.
Princeton University, Office of Communications, Chris Fascenell (2007)
According to Walczak, “No one has ever measured or shown how well reading out the concentration of these molecular gradients … actually pinpoints a specific position along the axis.”
Now they had: Even given the limited number of molecules and underlying noise of the system, the varying concentrations of the gap genes was sufficient to differentiate two neighboring cells in the head-to-tail axis — and the rest of the gene network seemed to be transmitting that information optimally.
“But the question always remained open: Does the biology actually care?” Gregor said. “Or is this just something that we measure?” Could the regulatory regions of DNA that responded to the gap genes really be wired up in such a way that they could decode the positional information those genes contained?
The biophysicists teamed up with the Nobel Prize-winning biologist Eric Wieschaus to test whether the cells were actually making use of the information potentially at their disposal. They created mutant embryos by modifying the gradients of morphogens in the very young fly embryos, which in turn altered the expression patterns of the gap genes and ultimately caused pair-rule stripes to shift, disappear, get duplicated or have fuzzy edges. Even so, the researchers found that their decoder could predict the changes in mutated pair-rule expression with surprising accuracy. “They show that the map is broken in mutants, but in a way that the decoder predicts,” Walczak said.
Lucy Reading-Ikkanda/Quanta Magazine
“You could imagine that if it was getting information from other sources, you couldn’t trick [the cells] like that,” Brewster added. “Your decoder would fail.”
These findings represent “a signpost,” according to Kondev, who was not involved with the study. They suggest that there’s “some physical reality” to the inferred decoder, he said. “Through evolution, these cells have figured out how to implement Bayes’ trick using regulatory DNA.”
How the cells do it remains a mystery. Right now, “the whole thing is kind of wonderful and magical,” said John Reinitz, a systems biologist at the University of Chicago.
Even so, the work provides a new way of thinking about early development, gene regulation and, perhaps, evolution in general.
A Steeper Landscape
The findings provide a fresh perspective on Waddington’s idea of a developmental landscape. According to Gregor, their work indicates that there’s no need for 20 questions or a gradual refinement of knowledge after all. The landscape “is steep from the beginning,” he said. All the information is already there.
“Natural selection [seems to be] pushing the system hard enough so that it … reaches a point where the cells are performing at the limit of what physics allows,” said Manuel Razo-Mejia, a graduate student at the California Institute of Technology.

Eric Wieschaus, a biologist at Princeton University, and two of his colleagues were awarded the Nobel Prize in 1995 for their discovery of the genetic mechanisms that regulate early development in fruit fly embryos.
Princeton University, Office of Communications, Denise Applewhite (2007)
It’s possible that the high performance in this case is a fluke: Since fruit fly embryos develop very quickly, perhaps in their case “evolution has found this optimal solution because of that pressure to do everything very rapidly,” said James Briscoe, a biologist at the Francis Crick Institute in London who did not participate in this study. To really cement whether this is something more general, then, researchers will have to test the decoder in other species, including those that develop more slowly.
Even so, these results set up intriguing new questions to ask about the often-enigmatic regulatory elements. Scientists don’t have a solid grasp of how regulatory DNA codes for the control of other genes’ activities. The team’s findings suggest that this involves an optimal Bayesian decoder, which allows the regulatory elements to respond to very subtle changes in combined gap gene expression. “We can ask the question, what is it about regulatory DNA that encodes the decoder?” Kondev said.
And “what about it makes it do this optimal decoding?” he added. “That’s a question we could not have asked before this study.”
“That’s really what this work sets up as the next challenge in the field,” Briscoe said. Besides, there may be many ways of implementing such a decoder at the molecular level, meaning that this idea could apply to other systems as well. In fact, hints of it have been uncovered in the development of the neural tube in vertebrates, the precursor of their central nervous system — which would call for a very different underlying mechanism.
Moreover, if these regulatory regions need to perform an optimal decoding function, that potentially limits how they can evolve — and in turn, how an entire organism can evolve. “We have this one example … which is the life that evolved on this planet,” Kondev said, and because of that, the important constraints on what life can be are unknown. Finding that cells show Bayesian behavior could be a hint that processing information effectively may be “a general principle that makes a bunch of atoms stuck together loosely behave like the thing that we think is life.”
But right now, it is still only a hint. Although it would be “kind of a physicist’s dream,” Gregor said, “we are far from really having proof for this.”
From Wires Under Oceans to Neurons in the Brain
The concept of information optimization is rooted in electrical engineering: Experts originally wanted to understand how best to encode and then decode sound to allow people to talk on the telephone via transoceanic cables. That goal later turned into a broader consideration of how to transmit information optimally through a channel. It wasn’t much of a leap to apply this framework to the brain’s sensory systems and how they measured, encoded and decoded inputs to produce a response.
Now some experts are trying to think about all kinds of “sensory systems” in this way: Razo-Mejia, for instance, has studied how optimally bacteria sense and process chemicals in their environment, and how that might affect their fitness. Meanwhile, Walczak and her colleagues have been asking what a “good decoding strategy” might look like in the adaptive immune system, which has to recognize and respond to a massive repertoire of intruders.
“I don’t think optimization is an aesthetic or philosophical idea. It’s a very concrete idea,” Bialek said. “Optimization principles have time and again pointed to interesting things to measure.” Whether or not they are correct, he considers them productive to think about.
“Of course, the difficulty is that in many other systems, the property being decoded is more difficult than one-dimensional position [along the embryo’s axis],” Walczak said. “The problem is harder to define.”
That’s what made the system Bialek and his colleagues studied so tantalizing. “There aren’t many examples in biology where a high-level idea, like information in this case, leads to a mathematical formula” that is then testable in experiments on living cells, Kondev said.
It’s this marriage of theory and experiment that excites Bialek. He hopes to see the approach continue to guide work in other contexts. “What’s not clear,” he said, “is whether the observation [of optimization] is a curiosity that arises in a few corners, or whether there’s something general about it.”
If the latter does prove to be the case, “then that’s very striking,” Briscoe said. “The ability for evolution to find these really efficient ways of doing things would be an incredible finding.”
Kondev agreed. “As a physicist, you hope that the phenomenon of life is not just about the specific chemistry and DNA and molecules that make living things on planet Earth — that it’s broader,” he said. “What is that broader thing? I don’t know. But maybe this is lifting a little bit of the veil off that mystery.”
Correction added on March 15: The text was updated to acknowledge the contributions of Mariela Petkova and Gašper Tkačik.
This article was reprinted on Wired.com.