Universal Method to Sort Complex Information Found

Eric Petersen for Quanta Magazine
Introduction
If you were opening a coffee shop, there’s a question you’d want answered: Where’s the next closest cafe? This information would help you understand your competition.
This scenario is an example of a type of problem widely studied in computer science called “nearest neighbor” search. It asks, given a data set and a new data point, which point in your existing data is closest to your new point? It’s a question that comes up in many everyday situations in areas such as genomics research, image searches and Spotify recommendations.
And unlike the coffee shop example, nearest neighbor questions are often very hard to answer. Over the past few decades, top minds in computer science have applied themselves to finding a better way to solve the problem. In particular, they’ve tried to address complications that arise because different data sets can use very different definitions of what it means for two points to be “close” to one another.
Now, a team of computer scientists has come up with a radically new way of solving nearest neighbor problems. In a pair of papers, five computer scientists have elaborated the first general-purpose method of solving nearest neighbor questions for complex data.
“This is the first result that captures a rich collection of spaces using a single algorithmic technique,” said Piotr Indyk, a computer scientist at the Massachusetts Institute of Technology and influential figure in the development of nearest neighbor search.
Distance Difference
We’re so thoroughly accustomed to one way of defining distance that it’s easy to miss that there could be others. We generally measure distance using “Euclidean” distance, which draws a straight line between two points. But there are situations in which other definitions of distance make more sense. For example, “Manhattan” distance forces you to make 90-degree turns, as if you were walking on a street grid. Using Manhattan distance, a point 5 miles away as the crow flies might require you to go across town for 3 miles and then uptown another 4 miles.
It’s also possible to think of distance in completely nongeographical terms. What is the distance between two people on Facebook, or two movies, or two genomes? In these examples, “distance” means how similar the two things are.
There exist dozens of distance metrics, each suited to a particular kind of problem. Take two genomes, for example. Biologists compare them using “edit distance.” Using edit distance, the distance between two genetic sequences is the number of additions, deletions, insertions and replacements required to convert one into the other.
Edit distance and Euclidean distance are two completely different notions of distance — there’s no way to reduce one to the other. This incommensurability is true for many pairs of distance metrics, and it poses a challenge for computer scientists trying to develop nearest neighbor algorithms. It means that an algorithm that works for one type of distance won’t work for another — that is, until this new way of searching came along.
Squaring the Circle
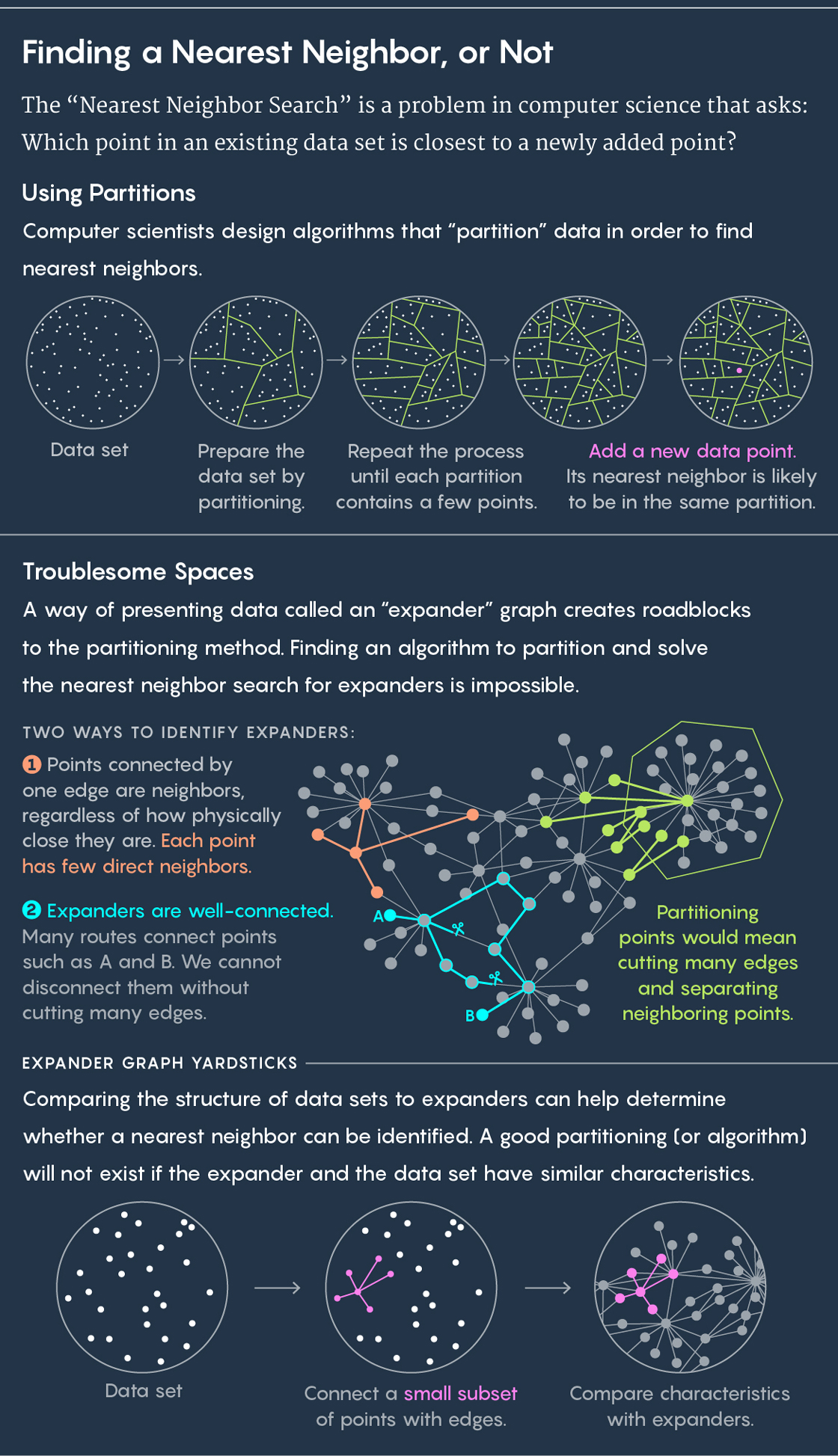
To find a nearest neighbor, the standard approach is to partition your existing data into subgroups. Imagine, for instance, your data is the location of cows in a pasture. Draw circles around groups of cows. Now place a new cow in the pasture and ask, which circle does it fall in? Chances are good — or even guaranteed — that your new cow’s nearest neighbor is also in that circle.
Then repeat the process. Partition your circle into subcircles, partition those partitions, and so on. Eventually, you’ll end up with a partition that contains just two points: an existing point and your new point. And that existing point is your new point’s nearest neighbor.
Lucy Reading-Ikkanda/Quanta Magazine
Algorithms draw these partitions, and good algorithm will draw them quickly and well — with “well” meaning that you’re not likely to end up in a situation where your new cow falls in one circle but its nearest neighbor stands in another. “From these partitions we want close points to end up in the same disc often and far points to end up in the same disc rarely,” said Ilya Razenshteyn, a computer scientist at Microsoft Research and co-author of the new work along with Alexandr Andoni of Columbia University, Assaf Naor of Princeton University, Aleksandar Nikolov of the University of Toronto and Erik Waingarten of Columbia University.
Over the years, computer scientists have come up with various algorithms for drawing these partitions. For low-dimensional data — where each point is defined by only a few values, like the locations of cows in a pasture — algorithms create what are called “Voronoi diagrams,” which solve the nearest neighbor question exactly.
For higher-dimensional data, where each point can be defined by hundreds or thousands of values, Voronoi diagrams become too computationally intensive. So instead, computer scientists draw partitions using a technique called “locality sensitive hashing (LSH)” that was first defined by Indyk and Rajeev Motwani in 1998. LSH algorithms draw partitions randomly. This makes them faster to run but also less accurate — instead of finding a point’s exact nearest neighbor, they guarantee you’ll find a point that’s within some fixed distance of the actual nearest neighbor. (You can think of this as being like Netflix giving you a movie recommendation that’s good enough, rather than the very best.)
Since the late 1990s, computer scientists have come up with LSH algorithms that give approximate solutions to the nearest neighbor problem for specific distance metrics. These algorithms have tended to be very specialized, meaning an algorithm developed for one distance metric couldn’t be applied to another.
“You could get a very efficient algorithm for Euclidean distance, or Manhattan distance, for some very specific important cases. But we didn’t have an algorithmic technique that worked on a large class of distances,” said Indyk.
Because algorithms developed for one distance metric couldn’t be used in another, computer scientists developed a workaround strategy. Through a process called “embedding,” they’d overlay a distance metric for which they didn’t have a good algorithm on a distance metric for which they did. But the fit between metrics was usually imprecise — a square peg in a round hole type of situation. In some cases, embeddings weren’t possible at all. What was needed instead was an all-purpose way of answering nearest neighbor questions.
A Surprise Result
In this new work, the computer scientists began by stepping back from the pursuit of specific nearest neighbor algorithms. Instead, they asked a broader question: What prevents a good nearest neighbor algorithm from existing for a distance metric?
The answer, they thought, had to do with a particularly troublesome setting in which to find nearest neighbors called an “expander graph.” An expander graph is a specific type of graph — a collection of points connected by edges. Graphs have their own distance metric. The distance between two points on a graph is the minimum number of lines you need to traverse to get from one point to the other. You could imagine a graph representing connections between people on Facebook, for example, where the distance between people is their degree of separation. (If Julianne Moore had a friend who had a friend who is friends with Kevin Bacon, then the Moore-Bacon distance would be 3.)
An expander graph is a special type of graph that has two seemingly contradictory properties: It’s well-connected, meaning you cannot disconnect points without cutting many edges. But at the same time, most points are connected to very few other points. As a result of this last trait, most points end up being far away from each other (because the low-connectivity means you have to take a long, circuitous route between most points).
This unique combination of features — well-connectedness, but with few edges overall — has the consequence that it’s impossible to perform fast nearest neighbor search on expander graphs. The reason it’s impossible is that any effort to partition points on an expander graph is likely to separate close points from each other.
“Any way to cut the points on an expander into two parts would be cutting many edges, splitting many close points,” said Waingarten, a co-author of the new work.


Alexandr Andoni (above), Ilya Razenshteyn and Erik Waingarten are coauthors of the new work.
MIT


Jake Knapp


Lucia Mosner
From left: Alexandr Andoni, Ilya Razenshteyn and Erik Waingarten are co-authors of the new work.
MIT, Jake Knapp, Lucia Mosner
In the summer of 2016, Andoni, Nikolov, Razenshteyn and Waingarten knew that good nearest neighbor algorithms were impossible for expander graphs. But what they really wanted to prove was that good nearest neighbor algorithms were also impossible for many other distance metrics — metrics where computer scientists had been stymied trying to find good algorithms.
Their strategy for proving that such algorithms were impossible was to find a way to embed an expander metric into these other distance metrics. By doing so, they could establish that these other metrics had unworkable expanderlike properties.
The four computer scientists went to Assaf Naor, a mathematician and computer scientist at Princeton University, whose previous work seemed well-suited to this question about expanders. They asked him to help prove that expanders embed into these various types of distances. Naor quickly came back with an answer, but it wasn’t the one they had been expecting.
“We asked Assaf for help with that statement, and he proved the opposite,” said Andoni.
Naor proved that expander graphs don’t embed into a large class of distance metrics called “normed spaces” (which include distances like Euclidean distance and Manhattan distance). With Naor’s proof as a foundation, the computer scientists followed this chain of logic: If expanders don’t embed into a distance metric, then a good partitioning must be possible (because, they proved, expanderlike properties were the only barrier to a good partitioning). Therefore, a good nearest neighbor algorithm must also be possible — even if computer scientists hadn’t been able to find it yet.
The five researchers — the first four, now joined by Naor — wrote up their results in a paper completed last November and posted online in April. The researchers followed that paper with a second one they completed earlier this year and posted online this month. In that paper, they use the information they had gained in the first paper to find fast nearest neighbor algorithms for normed spaces.
“The first paper showed there exists a way to take a metric and divide it into two, but it didn’t give a recipe for how to do this quickly,” said Waingarten. “In the second paper, we say there exists a way to split points, and, in addition, the split is this split and you can do it with fast algorithms.”
Altogether, the new papers recast nearest neighbor search for high-dimensional data in a general light for the first time. Instead of working up one-off algorithms for specific distances, computer scientists now have a one-size-fits-all approach to finding nearest neighbor algorithms.
“It’s a disciplined way of designing algorithms for nearest neighbor search in whatever metric space you care about,” said Waingarten.
Update: On August 14, the researchers posted online the second of their two papers. This article has been updated with links to the second paper.