Bacterial Clones Show Surprising Individuality

Adrian du Buisson for Quanta Magazine
Introduction
Massed at the starting line, the crowd of runners all looked identical. But this wasn’t your standard 5K. Instead, researchers wanted to test both speed and navigational ability as competitors wound their way through a maze, choosing the right direction at every intersection. At the end of the course, the postdocs Mehdi Salek and Francesco Carrara would be waiting to identify each of the finishers. The postdocs wouldn’t have any medals or a commemorative T-shirt for the winners, however, because their racers weren’t human. They were Escherichia coli bacteria.
That there could be individual winners at all is a notion that has shaken the foundations of microbiology in recent years. Working in the lab of Roman Stocker at the Swiss Federal Institute of Technology Zurich (ETH Zurich), a team of microbiologists and engineers invented this unique endurance event. The cells at the starting line of Stocker’s microbial marathon were genetically identical, which implied, according to decades of biological dogma, that their resulting physiology and behavior should also be more or less the same, as long as all the cells experienced identical environmental conditions. At the DNA level, every E. coli cell had a roughly equal encoded ability to swim and steer through the course. A pack of cells that started the race at the same time would in theory all finish around the same time.
But that’s not what Salek and Carrara found. Instead, some bacteria raced through the maze substantially more quickly than others, largely because of varying aptitude for moving toward higher concentrations of food, a process called chemotaxis. What appeared to Salek and Carrara as a mass of indistinguishable cells at the beginning was actually a conglomerate of unique individuals.
“Bacteria can be genetically identical but phenotypically different,” Carrara said.
This bacterial individuality — known more technically as phenotypic heterogeneity — upends decades of traditional thinking about microbes. Although scientists knew that, for example, antibiotics didn’t always kill every last microbe in a colony of identical clones, both the cause of these differences and the resulting implications remained shrouded in mystery. Now advances in microscopy and microfluidics (the technology Stocker’s lab used to build the bacterial maze) have begun to lift the veil on an important evolutionary process.
“This has been a relatively overlooked phenomenon,” said Hesper Rego, a microbiologist at the Yale School of Medicine. “The idea that microbial populations could evolve heterogeneity and control it using genetics is a really powerful concept.”
From Populations to Individuals
Ever since the days of Robert Koch and Louis Pasteur in the 1870s, microbiologists have typically studied groups of bacteria rather than individuals. Much of this was out of necessity: The technology didn’t exist to allow scientists to do much more with single cells than peer at them through a microscope. Besides, if the bacteria were all identical, then there seemingly wasn’t a need to study every cell. An individual cell deposited on a plate of nutrient-rich jelly would divide and divide until it formed a visible colony of cells, all clones of the original cell. All the bacteria in this colony could be expected to show the same behaviors, physiology and physical appearance — the same phenotype — when placed in identical environments. By and large, they did.
The development of antibiotics in the 1940s revealed a curious anomaly, however. In many cases, antibiotics didn’t annihilate all the bacteria, even in groups of cells that were fully susceptible to the killing power of antibiotics. The surviving cells were considered “persistent.” They just hunkered down and waited out the chemical barrage of penicillin or similar drugs. Initially, scientists thought that persisters might come from a genetically distinctive subpopulation that grew more slowly even before the antibiotic treatments. But when microbiologists looked for genes that could predict which cells would become persisters, they were disappointed.
“There was no such [distinct persistent] subpopulation,” said Laurence van Melderen, a microbiologist at the Free University of Brussels in Belgium. “In every population, you will find some persisters if you look for them.” For scientists, this posed a major quandary: How could identical bacteria have such radically different behaviors?
By the late 1970s, researchers had identified one possible answer. Scientists at the University of California, Berkeley showed that random chance alone could lead to different behaviors even in genetically identical cells. Bacteria with whiplike flagella can swim in a straight line (known as “running”) or lurch in random directions. Swimming cells spend much of their time tumbling about, actively sampling their environment. But to move toward higher concentrations of nutrients and away from toxins and predators, bacteria must use a direct run. When they can no longer sense a gradient, they return to tumbling.
Berkeley microbiologists studying E. coli found that each cell stopped swimming and started tumbling at a different concentration of various chemical attractants, including aspartate and L-serine. Even after considering random statistical variations and any influence from unlikely spontaneous mutations during the experiment, the researchers couldn’t account for the cells’ marked and persistent individual differences in running and tumbling. That mystery, according to Thierry Emonet, a biophysicist at Yale, was “a big deal.”
The study appeared during the heyday of the idea that a single gene made a single protein, which would subsequently elicit a consistent behavior when all the cells were in the same environment. After a century of experimentation on batches of bacteria, scientists were accustomed to slight collective deviations in “identical” traits, but their data still tended to cluster tightly around a mean. The Berkeley scientists, in contrast, found that sensitivity to the attractants was smeared out over a broad concentration, not a single mean. Their paper challenged the general assumption by showing substantial cell-to-cell variation in swimming behavior among the individual bacteria. No longer could phenotypic heterogeneity be shrugged off as a quirk of the bacterial response to antibiotics.
Although the researchers knew that this individuality resulted both from how tightly each cell regulated tumbling and from its response to L-serine, quantifying this variation in specific cells was more challenging. In 2002, glowing E. coli changed all of that.
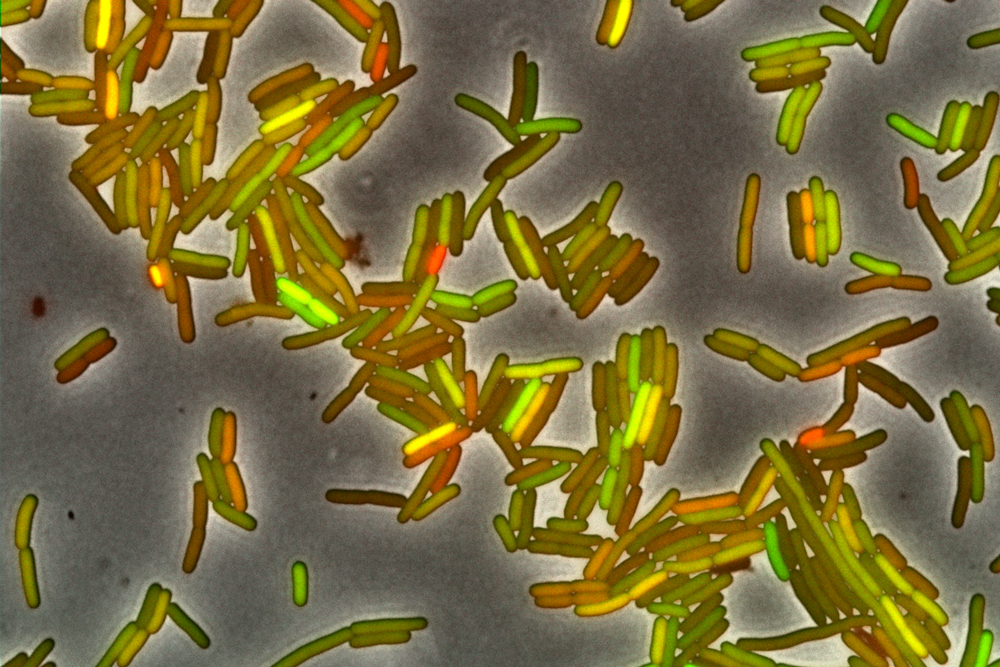
The cloned E. coli bacteria growing in this laboratory culture glow with colors from two fluorescent proteins they express. Their colors differ because even though the cells are genetically identical, they are functionally individuals: Stochastic noise in their gene expression makes them produce different amounts of the proteins.
The biophysicist Michael Elowitz, now at the California Institute of Technology, inserted two fluorescent genes — one yellow, one cyan — into specimens of E. coli. The fluorescent genes were under the control of the exact same machinery, so prevailing wisdom held that the bacteria would glow a uniform green, a constant mixture of the yellow and blue.
Yet they didn’t. Elowitz and his colleagues found that the ratio of yellow and cyan fluorescence varied from cell to cell, proving that gene expression varied among cells in the same environment. The team described that variation precisely in a 2002 Science paper. This work, van Melderen says, sparked a renaissance in the study of phenotypic heterogeneity.
Selection of Diversity
Advances in microscopy and microfluidics allowed researchers to build rapidly on Elowitz’s 2002 discovery. Two particular cellular behaviors — chemotaxis, or navigation along a chemical gradient, and the microbial stress response — figured prominently in their experiments. That’s because both of these responses, which are easily measured in a lab, allow cells to respond to a changing environment, according to Jessica Lee, a microbiology fellow at Global Viral who studied bacterial individuality as a postdoc in the lab of Chris Marx at the University of Idaho.
Take chemotaxis. If bacteria are moving toward something they like, they swim more and tumble less. But the point at which they make this switch varies from individual to individual, as Berkeley scientists discovered 40 years ago. Subsequent experiments revealed the existence of a family of chemotaxis proteins, such as one called CheY; the more copies of these proteins bacteria carried, the more likely they were to tumble instead of swim. Even without any environmental pressures affecting protein production, some bacteria may randomly have more molecules like CheY at any given time. Lee, Emonet and other researchers hypothesize that this innate variability lets a population of bacteria hedge its bets about the optimal amount of chemotaxis proteins for dealing with inevitable environmental changes.
Lee spent several years studying this bet-hedging behavior in the plant-dwelling bacterium Methylobacterium extorquens. Plants release oxygen as a byproduct of photosynthesis, but some plants also release methanol (wood alcohol). As its name suggests, M. extorquens can use this methanol as food, but the first step involves transforming the chemical into formaldehyde — the pungent chemical that works as a preservative because it is toxic to bacteria. M. extorquens bacteria protect themselves by breaking down the formaldehyde into a less toxic metabolite as quickly as possible. That’s how the bacteria are essentially able to “not pickle themselves,” Lee quipped.

In this time-lapse video of a cell culture, genetically identical Methylobacterium extorquens use methanol as a food source but produce toxic formaldehyde in the process. Because some of the cells exhibit a higher tolerance for the toxin, they can grow into colonies much more quickly.
Courtesy Jessica Audrey Lee
Because methanol isn’t always available and the metabolic machinery for thoroughly breaking down formaldehyde costs a lot of energy to produce, M. extorquens mostly doesn’t bother making the needed enzymes until the alcohol is actually present. But then the bacteria face a dilemma. When they start to break down methanol, the essential enzymes aren’t yet being produced at full capacity, so the soaring buildup of formaldehyde can kill the cells. Managing formaldehyde concentrations is life or death.
What Lee and Marx found, however, was that individual cells had different sensitivities to formaldehyde concentrations. As the scientists described in a paper posted earlier this year on the preprint server biorxiv.org, some bacteria continued to grow in the face of formaldehyde concentrations that killed most of their compatriots, even though all the cells were genetic clones.
“The only way we could explain it was that possibly the bacteria we thought were completely identical were in fact behaving in a not identical way,” Lee said. Something in the physiology of this formaldehyde-tolerant subpopulation — the scientists still don’t know what — allows it to survive and thrive in the presence of a deadly chemical. It’s the perfect example of a bet-hedging strategy, Lee says.
But this heterogeneity might have a significance that goes beyond improving the odds of survival for some members of a bacterial community. Scientists have also discovered hints that that bacterial individuality could have contributed to the evolution of multicellular organisms.
For example, experiments by the biophysicist Teun Vissers at the University of Edinburgh revealed that E. coli clones vary in their ability to stick to surfaces. The bet-hedging explanation for these differences is that because some cells may survive when others get washed away, the bacterial community as a whole benefits.
Yet the microbial ecologist Martin Ackermann at ETH Zurich highlights an additional hypothesis: His own work with Salmonella and other organisms has shown that when groups of identical cells diversify, they can divide up some of their tasks and start to specialize in certain processes.
“A benefit emerges through some interaction between the subpopulations. I think division of labor is a much more precise term” for the situation, Ackermann said. Evolutionary theorists often cite the division of labor and subsequent specialization of tasks among collections of single-celled organisms as a likely major factor driving the emergence of multicellularity.
The crucial question is: What is making these bacteria into distinct individuals if it isn’t their genetics? What is the source of this variation? Researchers are still searching for answers, but it is clear that this individuality isn’t simply the result of noise in the system. Random factors may figure into it, but specific mechanisms also somehow seem to be impressing cell-to-cell differences across bacterial populations.
Rego’s work on the tuberculosis bacterium Mycobacterium tuberculosis and a related species showed how some differences can arise during mitotic cell division. When a bacterium divides, it doesn’t produce two identical daughter cells. Instead, as the cell grows and elongates during the prelude to division, it must synthesize additional cellular material. Because this material tends to be concentrated on one side of the original cell, one daughter cell inherits newer parts than the other. This lopsidedness is especially pronounced in bacteria like M. tuberculosis. Rego was able to find a gene responsible for nearly all of this asymmetry, and when she manipulated it to make the two daughter cells more even, she eliminated nearly all the heterogeneity in the bacteria’s responses. This result suggests that the bacteria’s individuality is an adaptive advantage.
These recent advances in understanding the origins and functions of bacterial individuality still don’t completely explain the paradox that such nongenetic benefits can be maintained over billions of years of evolution. The secret to the maintenance of this heterogeneity, scientists suspect, is not in the traits themselves but rather in how these traits are regulated at the cellular level. Many genes essential to life are tightly controlled, since too little or too much activity means certain death. Natural selection may be indifferent to the regulation of other traits and may even allow for greater survival of populations that have higher variability. Phenotypic heterogeneity seems to fall into this second category. Having some organisms grow more slowly may seem to be a biological dead end, but if these same cells can weather an antibiotic storm, tolerance for a wider variation in growth rates may be a good thing. “In biology, you never have a single cell doing something. You have a group of cells,” Emonet said. “The diversity will affect the average performance of the group.”
Back in Zurich, in Salek and Carrara’s microbial racecourse, these advantages can be seen in those bacteria that race across the finish line and those that barely make it out of the starting gate. Far from being billions of identical clones, bacteria can display remarkable differences, even when they all share the same DNA. And it’s only by watching these microscopic dramas unfold over time that scientists have come to understand the diversity inherent in even the most identical populations.
“It’s changed our view of microorganisms,” van Melderen said. “Bacteria and other microorganisms are probably not as simple as we used to think. This phenotypic heterogeneity adds a level of complexity to every process.”