‘Broadband’ Networks of Viruses May Help Bacteria Evolve Faster
Viruses called bacteriophages (red) attack a bacterial cell in this artist’s conception. Scientists have discovered that such viruses can transfer genes between bacteria much more frequently than was thought — an arrangement that may benefit both the viruses and the bacteria.
Introduction
Bacteria have a sneaky evolutionary advantage: their own version of the internet for swapping survival solutions. It’s a living network of viruses that can shuttle genetic information between unrelated cells. Known as transduction, this process is one of the ways that bacteria can bypass the generation-by-generation plodding of vertical inheritance and instead share information horizontally, enabling genes slowly shaped by natural selection to enter a new population in an instant.
Scientists have known this transduction network must influence bacterial evolution across the sweep of centuries, but they presumed its short-term impact might be limited because transduction events seemed rare. A study published last week in Science, however, discovered a new mechanism of transduction that occurs 1,000 times more often, and that may accelerate bacterial evolution to a similar degree. Transduction may in fact be a central force in bacterial evolution.
“We had assumed that transduction occurred at rates analogous to dial-up internet,” said John Chen, an assistant professor of microbiology and immunology at the National University of Singapore and a lead co-author on this study. “But it appears that in some cases, transduction rates are more akin to broadband.” Their results not only suggest that transduction makes horizontal gene transfer much more common, but also that this bacterial internet might have been shaped by selection for the benefit of both bacteria and the viruses called bacteriophages.
A bacteriophage resembles a lunar landing module crossed with a spider, like something out of an arachnophobic astronaut’s nightmare. It lands on a bacterium, pierces the cell membrane and injects its DNA into the cell. From there, the virus hijacks the host’s cellular machinery to make copies of itself, which then bust out of the cell to infect other bacteria. On rare occasions, bits of bacterial DNA get packaged in the phage capsules instead and get added to the genomes of subsequently infected bacteria. This genetic transfer is known as general transduction.
A bacteriophage has another option, too: Its genes can slip into the bacterium’s genome unnoticed and lie in wait. Known as a prophage, this stretch of foreign DNA can persist for generations before activating. Approximately half of all sequenced bacteria contain a prophage, and many house more than one.
Once activated, the prophage cuts itself out of the bacterial genome, replicates and then packages its DNA into phages. Occasionally, there’s an error in the cutting, and a small portion of the bacterial genome gets copied and packaged alongside the prophage. When that bacteriophage infects another host, the bit of bacterial DNA gets incorporated into the new host’s genome in a process called specialized transduction.
Generalized transduction events are rare, happening on the order of once per million phages, and specialized transductions are even less common. Still, fluke occurrences can be beneficial on an evolutionary timescale. But the speed with which some bacteria adapt to environmental changes puzzled Chen. He studies Staphylococcus aureus, the notorious human pathogen behind staph infections that evolves resistance to antibiotics remarkably fast.
Resistant strains of staph almost always emerge within a year or two after the introduction of a new drug. Methicillin-resistant Staphylococcus aureus, or MRSA, is one well-known example: Resistant strains appeared soon after the antibiotic methicillin was introduced in the 1950s, and now MRSA is a scourge in hospitals worldwide. “[S. aureus] adapts extremely rapidly, and I felt there was something more to it,” Chen said. The known transduction-mechanism explanations weren’t satisfying to him, “but that was just a gut feeling, not a scientific conclusion.”
Chen’s suspicions deepened after seeing a curious result from his collaborator José Penadés of the University of Glasgow, a co-author on this study. When Penadés and Chen looked at the progression of phage production in S. aureus, they noticed an odd discrepancy: It seemed like the phage DNA was being replicated and packaged long before any viral DNA was being excised from the host genome.
This delay had potentially profound implications. If the prophage DNA was getting replicated and stuffed into viral capsules while still embedded in the host genome, then there would be many more opportunities for slip-ups in which adjacent chunks of bacterial DNA would also get copied and encased in a phage — immensely expanding the chances for a horizontal transfer of genes to other bacterial species.
This delayed-excision mechanism had been seen in artificially generated mutants but never in nature. Chen, Penadés and their coworkers demonstrated not only that this new mechanism of transduction occurs naturally in S. aureus, but also that it is 1,000-fold as frequent as other modes of transduction.
“We were blown away,” Chen said. “We were expecting relatively low levels, but this was extreme.” He added, “We showed the data to some of our colleagues, and they were practically in disbelief.”
“Lateral transduction,” as the researchers called it, also captures larger swaths of the genome than general and specialized transduction do, especially for bacteria with multiple prophages within their genome. “We now understand that much larger blocks of the genome are hypermobile,” Chen said.
“These results suggest transduction is a much stronger force in bacterial evolution than we’d previously realized,” said Alan Davidson, a professor of microbiology at the University of Toronto who was not involved in this study. “It could really help us explain not just long-term evolution, but short-term evolution as well.”
Davidson also noted that some unexplained quirks of genomic data make more sense in light of lateral transduction. He cites a recent study in which mice were treated with ciprofloxacin, an antibiotic that works by activating prophages. The treatment produced a bloom of antibiotic- resistance genes among the phage viruses in the mice, consistent with prophage activation causing a surge in lateral transduction.
Another mystery that lateral transduction could solve is the existence of “pathogenicity islands” — chunks of bacterial genomes that contain a genetic toolkit for evading antibiotic resistance and becoming more virulent. They dot the genomes of many pathogenic species of Staphylococcus, but are oddly absent in their nonpathogenic close relatives. Scientists struggled to explain this pattern, but lateral transduction provides a plausible mechanism.
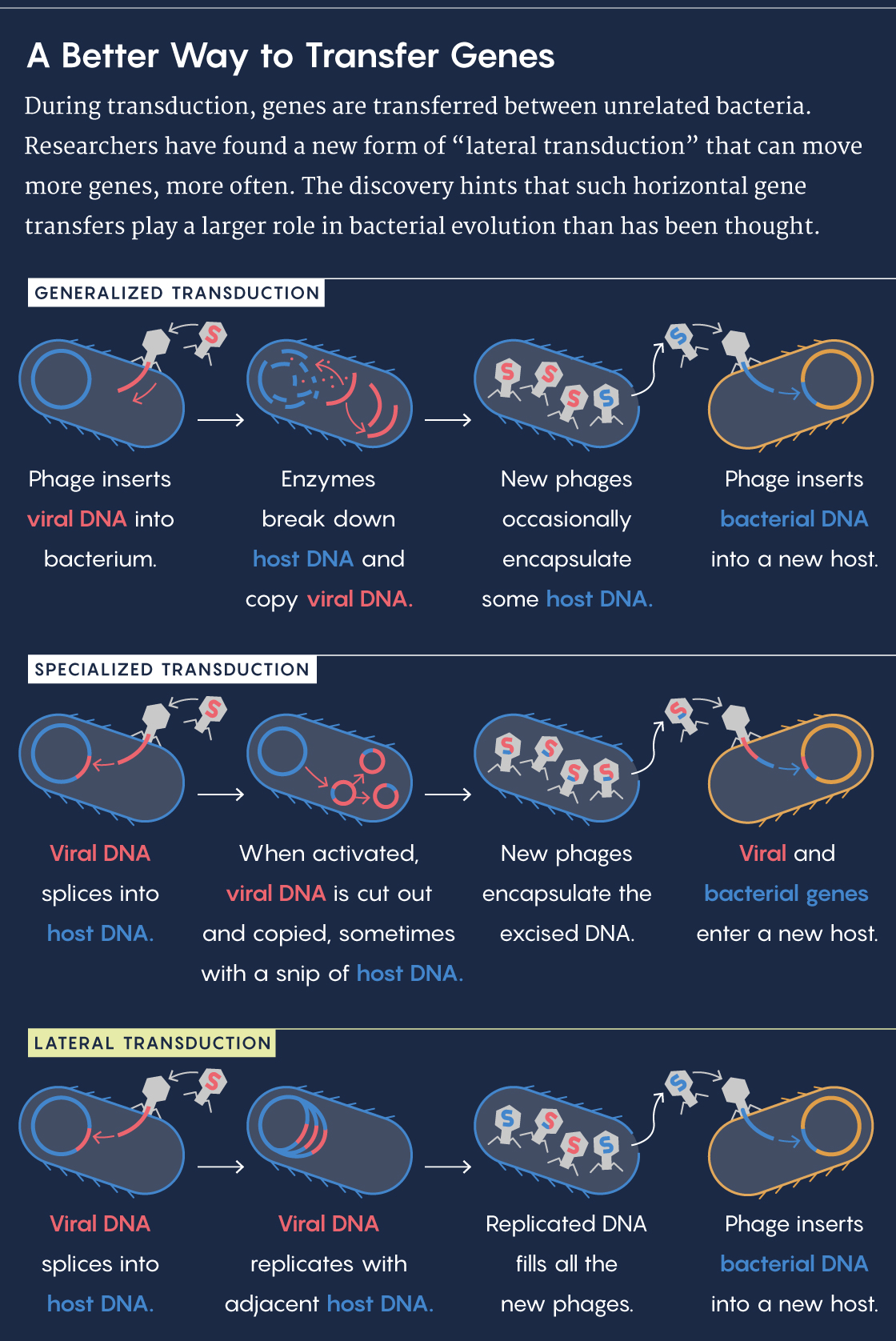
Lucy Reading-Ikkanda/Quanta Magazine
Indeed, although more work is needed to confirm it, some evidence hints that many S. aureus pathogenicity islands might cluster around prophage attachment sites. “It’s a very clever place to be located if you want to travel around the world,” Penadés said.
Penadés’s point raises the possibility that lateral transduction might be more of an innovation than a mistake, the product of a long co-evolutionary negotiation between bacteria and host. The virus might more easily spread by boosting the fitness and virulence of its host, while for the bacteria, the advantages of being plugged into the viral network of beneficial genes may outweigh the collateral damage from the prophage killing some cells.
Olga Zhaxybayeva, an associate professor at Dartmouth College who studies the co-evolution of bacteria and viruses and was not involved in this study, is intrigued by these findings. But she cautions against assuming selection has fine-tuned this mechanism for the benefit of both parties. “It’s definitely an interesting possibility and worth testing, but we can’t say for sure quite yet,” she said.
“These results are pretty amazing,” said Christine Schneider, a virologist and associate professor of biology at Carroll University. “It’s such an intriguing mechanism, and I’m curious to know if it’s widespread among other bacteria.”
Chen agrees that lateral transduction must be researched in more bacterial groups, but thinks the mechanism is likely widespread. His group plans to investigate more bacterial species and look for evidence that lateral transduction occurs in natural environments. “Lateral transduction, if common, could be really powerful,” Chen said. “A complete set of genes can be delivered to a bacterial cell that’s never seen it before, bypassing millions or even a billion years of evolution in a second.”