Giant Genetic Map Shows Life’s Hidden Links
Marta Iwanek for Quanta Magazine
Introduction
It’s a curious fact of biology: In yeast, only one in five genes is essential. If any of the approximately 1,200 critical genes are destroyed (out of 6,000), the result is death. Remove one of the others, and the yeast soldiers on.
The same is not always true, however, if a pair of nonessential genes is removed — sometimes, death comes quickly. In these cases, it’s likely that the genes have similar roles. They might both take out the cell’s garbage, for instance, or fix damaged DNA. The loss of one might not be deadly — the other could pick up the slack. But the loss of both is catastrophic.
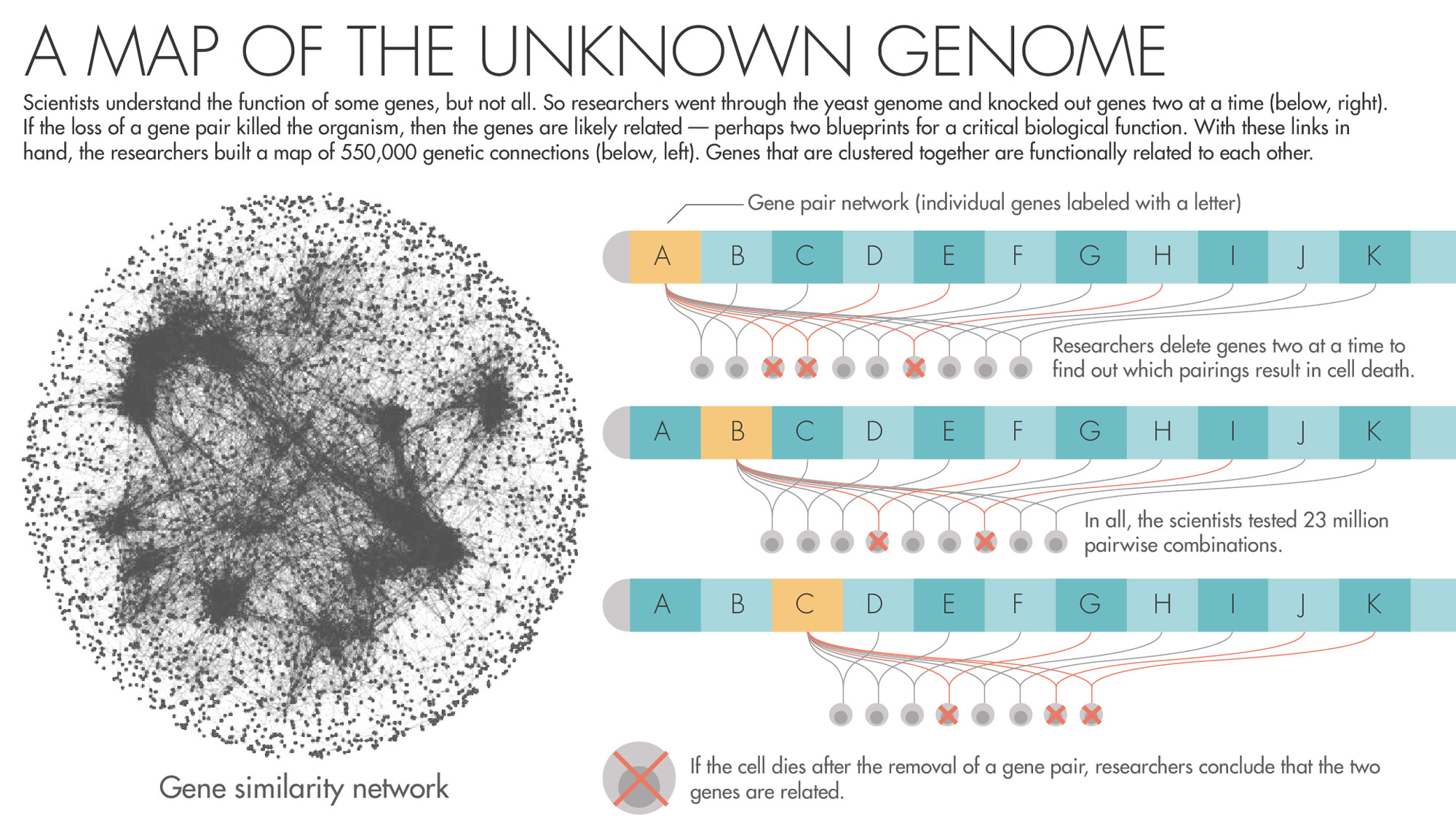
Can we use what happens when a pair of genes is destroyed to find out their function? This is the question that Charles Boone and Brenda Andrews, biologists at the University of Toronto, began to ask themselves about 17 years ago. If you know what one gene is doing in the cell, and destroying it kills the cell only if another, more mysterious gene goes too — can that give you clues to what the mystery gene does?
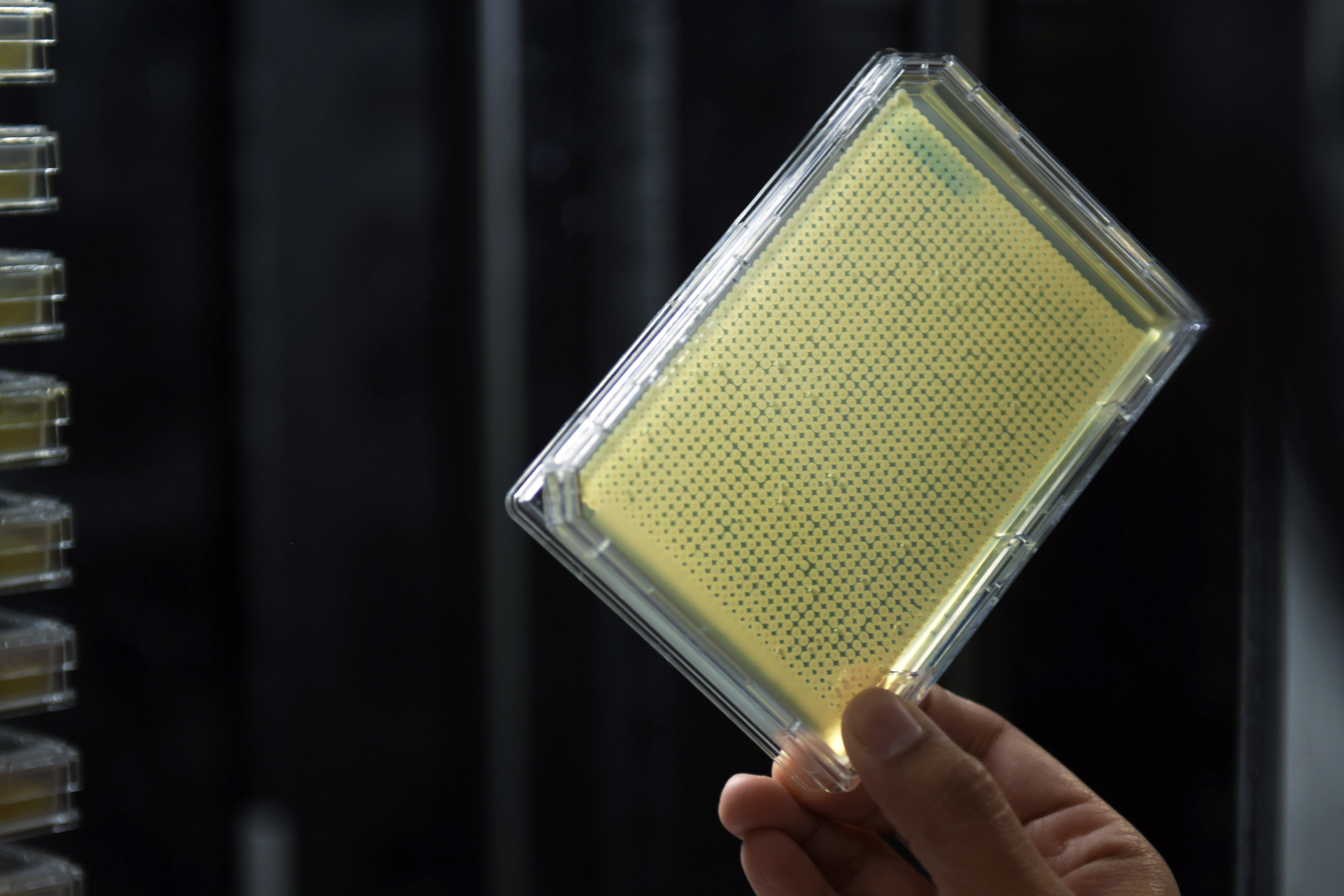
To answer the question, they began to orchestrate a precise campaign to destroy, two by two, all the genes in yeast. Using a fleet of yeast-growing robots, they created approximately 23 million strains of yeast, each effectively missing a pair of genes. By watching to see whether the yeast lived, died or grew sickly, the researchers generated data about the existence of relationships between the genes.

The Cell’s Backup Genetic Instructions
Now Boone, Andrews and a large team of collaborators have published in Science a sprawling report on the nearly two-decade-long set of experiments. In all, they found 550,000 pairs that, when removed, result in sickness or death. This network of genetic connections reveals a previously hidden scaffolding that underlies the operation of the cell. “The complete picture,” Boone said, “clearly shows a beautiful hierarchical structure.”
Over here are the genes involved in taking out the cell’s garbage, and over there are the genes responsible for its metabolism. Zoom out from one cluster of genes, and you’ll find the ones involved in the larger process the cluster is nested in. Zoom out from those and you’ll find all the ones that function alongside them in the same compartment of the cell. There’s something vertiginous in this view of life, a feeling that all the layers of complexity that let the organism thrive are there to look through, just as they were laid down by evolution.
As beautiful as the bird’s-eye view of the cell is, this work goes beyond biological voyeurism. This information can tell us about the evolution of the cell and, potentially, about how things go wrong in disease.

Yeast samples are put into a robotic device designed to process the experiment’s huge numbers of yeast strains.
Marta Iwanek for Quanta Magazine
Magisterial Maps
Using maps of interactions between genes or proteins is a popular approach to understanding the cell these days. Many researchers, looking at organisms from yeast to worms to humans, are building networks made up of proteins that attach to each other or genes that regulate each other. But the scale of Boone and Andrews’ effort sets it apart. In addition, their method can uncover connections that can’t be made by other tests, like those that focus on proteins that physically attach to one another. “It’s really a magisterial undertaking,” said David Botstein, the chief scientific officer of Google’s anti-aging startup Calico and a pioneer of genome mapping. When Boone and Andrews’ goal of knocking out all possible pairs of genes was floated years ago, “people thought, well, it’s just insane!” recalled Marian Walhout, a systems biologist at the University of Massachusetts Medical School. Even today, with advances in technology, it’s breathtaking, she said.
With the new information and the website where it can be navigated, researchers will be able to look up the genes they study and perhaps find that they have connections that have never been noticed before. “That utility is going to be, I predict, one of the major uses of the paper,” Botstein said. Earlier this month the yeast biochemist Yoshinori Ohsumi was awarded the Nobel Prize in physiology or medicine for his work on autophagy, the programmed destruction of pieces of the cell. “If he were doing his work now, he could go look at this data and see which genes genetically interact with the autophagy genes, and make huge progress much more quickly,” Walhout said.
For those of us who are not scientists, the research also provides an interesting reminder that the cell is not as simple as it might seem. Just because a gene is not one of the “essential” 1,200 does not mean it’s unimportant. It simply means that evolution has built many overlapping systems into the cell so that if one part goes, the whole thing doesn’t fall apart.
For example, the cell has a couple of different pathways that it can use to repair DNA. “Neither one is essential,” said Chad Myers, a computational biologist at the University of Minnesota who is a lead author of the new paper. But if you knock a single gene out in both of those pathways at the same time, “you’ll kill the cell,” he said. With both of those options for DNA repair gone, the cell can’t keep going.
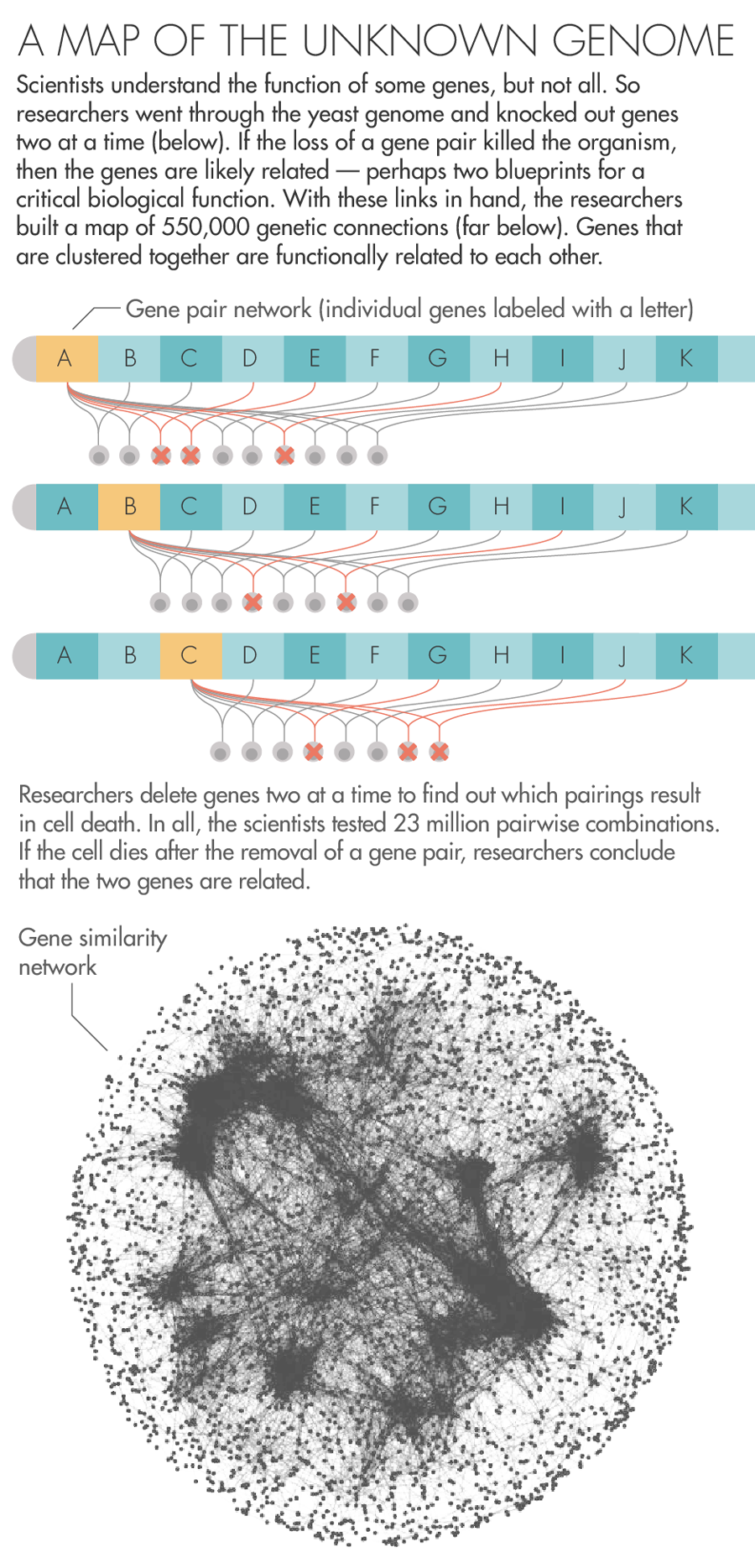
Lucy Reading-Ikkanda for Quanta Magazine; Gene similarity network courtesy of University of Toronto and University of Minnesota
Additionally, it’s worth remembering that lab-grown yeast live a life of relative luxury, with plenty of glucose to eat, a comfortable temperature, and so on. The pairs of genes whose removal caused the death of these yeast were needed to keep them alive under these very specific circumstances. But, Botstein said, “flexibility is the issue in survival — the ability to switch between circumstances that are good, or bad, or whatever, and not die.” Evolution has honed yeast to be ready for many kinds of situations, from the warm surface of a Napa Valley grape to the desert where a bird, having consumed the grape, might deposit it inside its droppings. If the experiments were repeated in myriad other circumstances, the removal of other combinations of genes would be deadly. Those experiments would reveal overlapping groups of genes that are essential for each of these situations, intimating the immense permutations that life is prepared for.
On a more immediate level, the yeast work has implications for human cells, which share many of the same biological mechanisms. Errors in multiple genes most likely underlie hereditary diseases that cannot be pinpointed to a single causal gene, as Walhout observes. People with an illness might have constellations of mutations in related pathways, and researchers are only beginning to untangle such cases. The yeast experiments may help suggest where the causes of human disorders lie. And they also make clear that the cell is a system where a handful of small changes can add up to a problem, though none of the individual alterations are harmful in and of themselves.
The team’s information has already begun to influence the direction of basic research. In 2013, Claire Moore, a biochemist at Tufts University, received an email from Boone. Moore uses yeast in her lab because it’s handy and extremely well-studied — so much so, in fact, that as of 2013 the functions of all but six of the essential genes were known — and she focuses on a process called polyadenylation, in which molecules are added to RNA to help get it to its destination in the cell.
Moore was vaguely aware of Boone’s work, but didn’t know him personally. “I get this email out of the blue,” she recalled. It said that Boone’s team had identified a gene that might be involved in polyadenylation. Could she check it out for them? “That was a surprise to me,” she said, “because I thought that we had identified all the proteins that did this.”
But Boone, Andrews and their collaborators had found that this gene, one of the essential, still-mysterious six, had very strong connections with those involved in polyadenylation. It was a simple matter for Moore to see whether knocking it out would interfere with the process in her yeast. And, lo and behold, it did. They had identified the mysterious gene’s function, merely by looking at a database of its relationships, and seeing what happened in the lab.
Moore, whose findings about the new polyadenylation gene appear in the Science paper, said that she thinks her experience shows the value of the network approach. “For us it’s opened up a whole new direction for the lab,” she said. “I don’t think we would have ever stumbled across it [otherwise].” She named the gene IPA1, which stands for “important for polyadenylation.” (Of her colleagues, she said with a chuckle, “they like that, because they like beer.”)
The Toronto researchers are already thinking about another big project: knocking out trios of genes instead of pairs. Many of their cells with two genes missing did not show any particular change from normal. But with a third gene removed, more cells will fail. Even if the groups test only a targeted set of genes to start out with, the group could uncover potentially thousands of new interactions. All in a day’s work — or a couple decades’.
This article was reprinted on TheAtlantic.com.