How Cells Pack Tangled DNA Into Neat Chromosomes
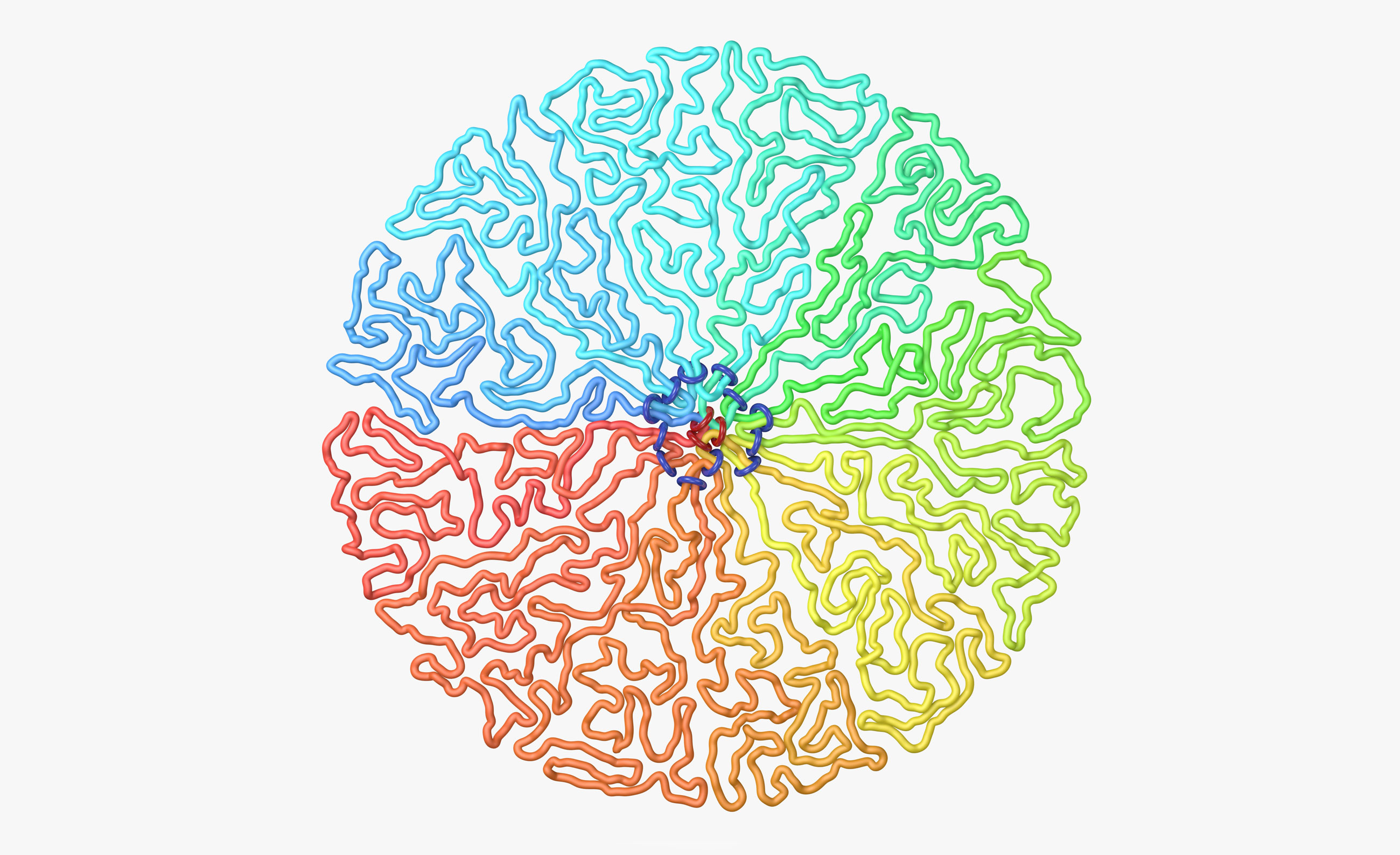
An illustrated cross-section of a dividing cell’s chromosome shows ring-shaped protein molecules folding the DNA into nested loops that radiate out from a central axis.
Dr. Anton Goloborodko
Introduction
A human cell carries in its nucleus two meters of spiraling DNA, split up among the 46 slender, double-helical molecules that are its chromosomes. Most of the time, that DNA looks like a tangled ball of yarn — diffuse, disordered, chaotic. But that messiness poses a problem during mitosis, when the cell has to make a copy of its genetic material and divide in two. In preparation, it tidies up by packing the DNA into dense, sausagelike rods, the chromosomes’ most familiar form. Scientists have watched that process through a microscope for decades: The DNA condenses and organizes into discrete units that gradually shorten and widen. But how the genome gets folded inside that structure — it’s clear that it doesn’t simply contract — has remained a mystery. “It’s really at the heart of genetics,” said Job Dekker, a biochemist at the University of Massachusetts Medical School, “a fundamental aspect of heredity that’s always been such a great puzzle.”
To solve that puzzle, Dekker teamed up with Leonid Mirny, a biophysicist at the Massachusetts Institute of Technology, and William Earnshaw, a biologist at the University of Edinburgh in Scotland. They and their colleagues used a combination of imaging, modeling and genomic techniques to understand how the condensed chromosome forms during cell division. Their results, published recently in Science and confirmed in part by experimental evidence reported by a European team in today’s issue of the journal, paint a picture in which two protein complexes sequentially organize the DNA into tight arrays of loops along a helical spine.
The researchers collected minute-by-minute data on chromosomes — using a microscope to see how they changed, as well as a technology called Hi-C, which provides a map of how frequently pairs of sequences in the genome interact with one another. They then generated sophisticated computer simulations to match that data, allowing them to calculate the three-dimensional path the chromosomes traced as they condensed.
Their models determined that in the lead-up to mitosis, a ring-shaped protein molecule called condensin II, composed of two connected motors, lands on the DNA. Each of its motors move in opposite directions along the strand while remaining attached to one another, causing a loop to form; as the motors continue to move, that loop gets larger and larger. (Mirny demonstrated the process for me by clasping a piece of his computer’s power cord with both hands, held knuckles to knuckles, through which he then proceeded to push a loop of cord.) As tens of thousands of these protein molecules do their work, a series of loops emerges. The ringlike proteins, positioned at the base of each loop, create a central scaffolding from which the loops emanate, and the entire chromosome becomes shorter and stiffer.
Those results lent support to the idea of loop extrusion, a prior proposal about how DNA is packaged. (Loop extrusion is also responsible for preventing duplicated chromosomes from becoming knotted and entangled, according to Mirny. The mechanics of the looped structure cause sister chromatids to repel each other.) But what the scientists observed next came as more of a surprise and allowed them to build further detail into the loop extrusion hypothesis.
After about 10 minutes, the nuclear envelope keeping the chromosomes together broke down, giving a second ring-shaped motor protein, condensin I, access to the DNA. Those molecules performed loop extrusion on the loops that had already formed, splitting each into around five smaller loops on average. Nesting loops in this way enabled the chromosome to become narrower and prevented the initial loops from growing large enough to mix or interact.
After approximately 15 minutes, as these loops were forming, the Hi-C data showed something that the researchers found even more unexpected. Typically, sequences located close together along the string of DNA were most likely to interact, while those farther apart were less likely to do so. But the team’s measurements showed that “things [then] kind of came back again in a circle,” Mirny said. That is, once the distance between sequences had grown even further, they again had a higher probability of interacting. “It was obvious from the first glance at this data that we’d never seen something like this before,” he said. His model suggested that condensin II molecules assembled into a helical scaffold, as in the famous Leonardo staircase found in the Chambord Castle in France. The nested loops of DNA radiated out like steps from that spiraling scaffold, packing snuggly into the cylindrical configuration that characterizes the chromosome.
“So this single process immediately solves three problems,” Mirny said. “It creates a scaffold. It linearly orders the chromosome. And it compacts it in such a way that it becomes an elongated object.”
“That was really surprising to us,” Dekker said — not only because they’d never observed the rotation of loops along a helical axis, but because the finding taps into a more fundamental debate. Namely, are chromosomes just a series of loops, or do they spiral? And if they do spiral, is it that the entire chromosome twists into a coil, or that only the internal scaffolding does? (The new study points to the latter; the researchers attribute the former helix-related hypothesis to experimental artifacts, the result of isolating chromosomes in a way that promoted excessive spiraling.) “Our work unifies many, many observations that people have collected over the years,” Dekker said.
“This [analysis] provides a revolutionary degree of clarity,” said Nancy Kleckner, a molecular biologist at Harvard University. “It takes us into another era of understanding how chromosomes are organized at these late stages.”

This series of images illustrates how a compacted chromosome takes shape. Ring-shaped motor proteins (red) form a helical scaffold. Folded loops of DNA emanate from that spiraling axis so that they can be packed tightly into a cylindrical rod.
Dr. Anton Goloborodko
Other experts in the field found those results less surprising, instead deeming the study more noteworthy for the details it provided. Hints of the general chromosomal assembly the researchers described were already “in the air,” according to Julien Mozziconacci, a biophysicist at Sorbonne University in France. The more novel aspects of the work, he said, lay in the researchers’ collection of Hi-C data as a function of time, which allowed them to pinpoint specific constraints, such as the sizes of the loops and helical turns. “I think this is a technical tour de force that allows us to see for the first time what people have been thinking,” he said.
Still, Dekker cautioned that, although it’s been known for some time that condensins are involved in this process — and despite the fact that his group has now identified more specific roles for those “molecular hands that cells use to fold chromosomes” — scientists still don’t understand exactly how they do it.
“If condensin is organizing mitotic chromosomes in this manner, how does it do so?” said Kim Nasmyth, a biochemist at the University of Oxford and a pioneer of the loop extrusion hypothesis. “Until we know the molecular mechanism, we can’t say for sure whether condensin is indeed the one driving all this.”
That’s where Christian Häring, a biochemist at the European Molecular Biology Laboratory in Germany, and Cees Dekker, a biophysicist (unrelated to Job Dekker) at Delft University of Technology in the Netherlands, enter the picture. Last year, they and their colleagues directly demonstrated for the first time that condensin does move along DNA in a test tube — a prerequisite for loop extrusion to be true. And in today’s issue of Science, they reported witnessing an isolated condensin molecule extruding a loop of DNA in yeast, in real time. “We finally have visual proof of this happening,” Häring said.
And it happened almost exactly as Mirny and his team predicted it would for the formation of their larger loops — except that in the in vitro experiment, the loops formed asymmetrically: The condensin landed on the DNA and reeled it in from only one side, rather than in both directions as Mirny initially assumed. (Since the experiments involved condensin from yeast, and only examined a single molecule at a time, they could neither confirm nor refute the other aspects of Mirny’s models, namely the nested loops and helical scaffold.)
Once researchers have completely unpacked that biochemistry — and conducted similar studies on how chromosomes unwind themselves — Job Dekker and Mirny think their work can lend itself to a range of practical and theoretical applications. For one, the research could inform potential cancer treatments. Cancer cells divide quickly and frequently, “so anything we know about that process can help specifically target those kinds of cells,” Dekker said.
It could also provide a window into what goes on in the chromosomes of cells that aren’t dividing. “It has wider implications for, I believe, any other thing the cell does with chromosomes,” Job Dekker said. The condensins he and his colleagues are studying have close relatives, called cohesins, that help with organizing the genome and creating loops even when the DNA isn’t getting compacted. That folding process could affect gene expression. Loop extrusion basically brings pairs of loci together, however briefly, at the base of the growing or shrinking loop — something that could very well be happening during gene regulation, when a gene has to be in physical contact with a regulatory element that may be located quite a distance away along the chromosome. “We now have such a powerful system to study this process,” Dekker said.
“I think there’s an incredible amount of synergy between the things we can learn at different parts of the cell cycle,” added Geoff Fudenberg, a postdoctoral researcher at the University of California, San Francisco, who previously worked in Mirny’s lab. Understanding how chromosomes undergo such a “dramatic transition” during mitosis, he said, could also reveal a lot about what they are doing “below the surface” when cells are not dividing and certain activities and behaviors are less clear.
Mirny points out that this type of folding could also provide insights into other processes in cells that involve active changes in shape or structure. Proteins get folded largely by interactions, while motor processes create the cytoskeleton in the cytoplasm. “Now we came to realize that chromosomes may be something in between,” Mirny said. “We need to gain a better understanding of how these types of active systems self-organize to create complex patterns and vital structures.”
Before that’s possible, the researchers will have to confirm and flesh out the solution they’ve proposed to what Job Dekker called a “great puzzle.” Kleckner has high hopes as well. “This work sets the foundation for a whole new way of thinking about what might be going on,” she said.
This article was reprinted on Wired.com.