How Heat Kills Cells
Introduction
Above a certain temperature, a cell will collapse and die. One of the most straightforward explanations for this lack of heat hardiness is that the proteins essential to life — the ones that extract energy from food or sunlight, fend off invaders, destroy waste products and so on — often have beautifully precise shapes. They start as long strands, then fold into helixes, hairpins and other configurations, as dictated by the sequence of their components. These shapes play a huge role in what they do. Yet when things start to heat up, the bonds that keep protein structures together break: first the weaker ones, and then, as the temperature mounts, the stronger ones. It makes sense that a pervasive loss of protein structure would be lethal, but until recently, the details of how, or if, this kills overheated cells were unknown.
Now, however, in a true tour de force, biophysicists at ETH Zurich in Switzerland have examined the behavior of every protein in cells from four different organisms as heat increases. This study and its rich deposit of data, published recently in Science, reveal that at the temperature at which a cell dies — whether it’s a human cell or one from Escherichia coli — only a handful of key proteins fall apart. Moreover, a protein’s abundance in a cell seems to show an intriguing relationship to the protein’s stability. The studies offer a glimpse into the fundamental rules that govern the order and disorder of proteins — rules that, researchers are realizing, have implications far beyond the question of why heat kills.
Paola Picotti, the biophysicist who led the study, explained that the experiments sprang from an old, thorny question: Why do some cells survive at high temperatures while others die? The bacterium Thermus thermophilus lives happily in hot springs and even in household hot water heaters, while E. coli withers above 40 degrees Celsius (104 degrees Fahrenheit). Strong evidence implies that differences in the stability of each organism’s proteins are involved. But looking at a protein’s behavior while it is still sitting in its living cell — the ideal way to understand it — is not easy. And isolating a protein in a test tube gives only partial answers, because within the organism, proteins nestle together, altering each other’s chemistry or holding each other in the right shape. To understand what is falling apart and why, you need to look at the proteins while they are still influencing each other.
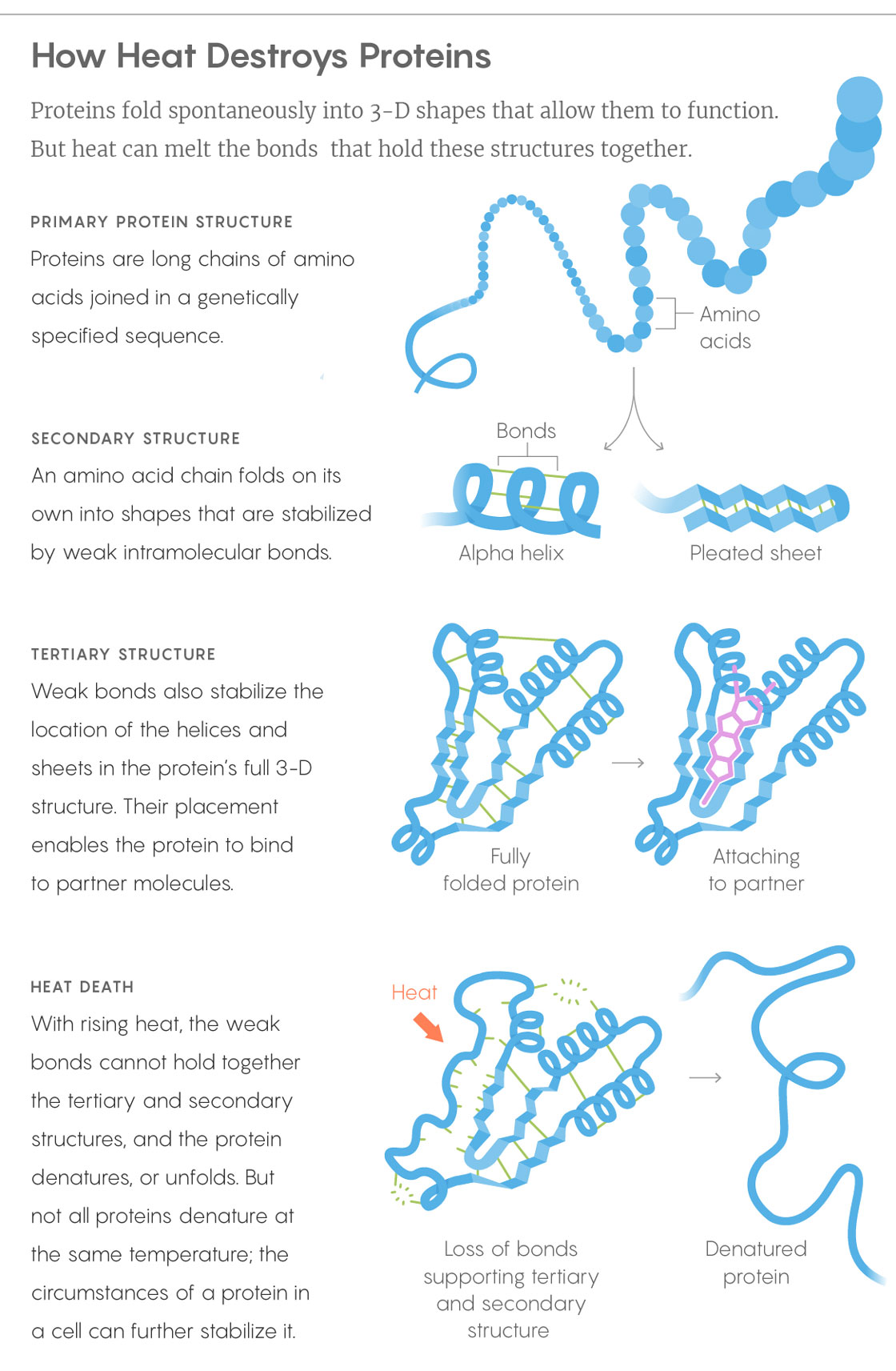
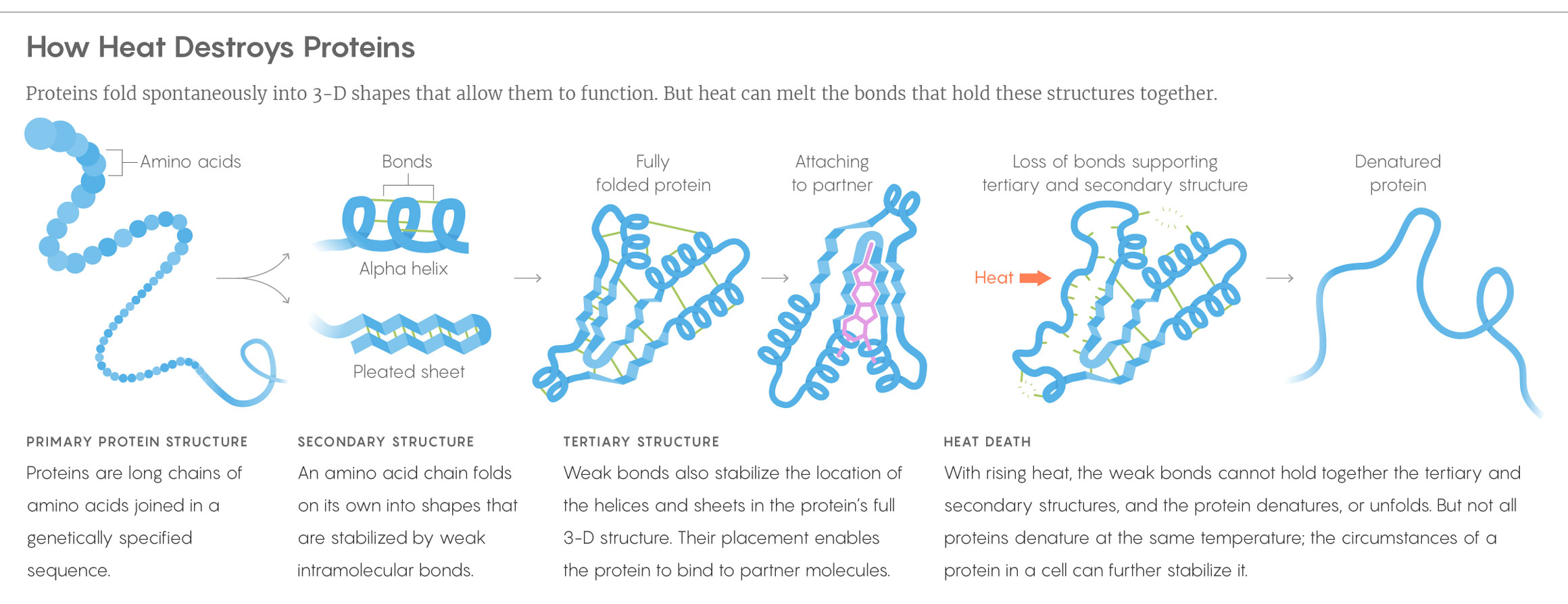
Lucy Reading-Ikkanda/Quanta Magazine
To address this problem, the team devised a sprawling automated workflow in which they split open cells and heated up their contents in stages, unleashing protein-slicing enzymes on the mixtures at every stage. These enzymes are particularly good at slicing up proteins that have unfolded, so the researchers could tell by looking at the fragments which proteins fell apart at each temperature hike. In this way, they graphed an unfolding, or denaturing, curve for each of the thousands of proteins they studied, showing its arc as it moved from an intact structure at comfortable temperatures to a denatured state as the degrees ticked up. To see how these curves differed across species, they performed the process on cells from four species — humans, E. coli, T. thermophilus and yeast. “This is a beautiful study,” said Allan Drummond, a biologist at the University of Chicago, emphasizing both the scale and the delicacy of the process.
One of the clearest observations was that in each species, the proteins did not unfold en masse with a temperature boost. Instead, “we saw that only a small subset of proteins collapses very early,” Picotti said, “and these are key proteins.” In a network-style diagram of proteins’ interrelations, these fragile few are often highly connected, meaning that they influence numerous processes in the cell. “Without these the cell cannot function,” Picotti said. “When these are gone, the whole network most likely collapses.” And with it, evidently, the life of the cell.
This paradox — that some of the most important proteins seem to be the most delicate — may reflect how evolution has shaped them to do their jobs. If a protein has many roles to play, it might gain an advantage from being somewhat unstable and prone to unfolding and refolding, since this could allow it to assume various shapes appropriate to whatever its next target might be. “Many of these [key] proteins have high flexibility, which makes them more unstable,” but it may give them the versatility to bind to a variety of target molecules in the cell, Picotti explained. “That’s how they can perform their function, most likely. … It’s a trade-off.”
Looking more closely at E. coli, for which they had the cleanest data, the researchers also found a relationship between a protein’s abundance — how many copies of it are floating around the cell — and its stability. The more copies the cell made, they reported, the more heat it took to break a protein down. (Abundance, it should be noted, doesn’t necessarily correlate with being essential for life: some rare proteins are crucial.) This connection between abundance and sturdiness supports an idea that Drummond put forward a decade or so ago, concerning the cellular protein-making machinery’s tendency to make occasional errors. A mistake usually destabilizes a protein. If that protein happens to be a common one, produced by the hundreds or thousands in a cell daily, then misfolded copies made in large numbers could fatally clog the cell. It would behoove an organism to evolve versions of common proteins with extra stability built in, and the Picotti team’s data seem to reflect this.
To explore what qualities make a protein heat stable, the researchers compared the data from E. coli and T. thermophilus. E. coli proteins began to fall apart at 40 degrees Celsius and had mostly degraded by 70 degrees Celsius. But at that temperature, T. thermophilus proteins were just starting to get uncomfortable: Some of them continued to hold their shape up to at least 90 degrees Celsius. The team found that the T. thermophilus proteins tended to be shorter, and certain kinds of shapes and components cropped up more often in the stablest proteins.
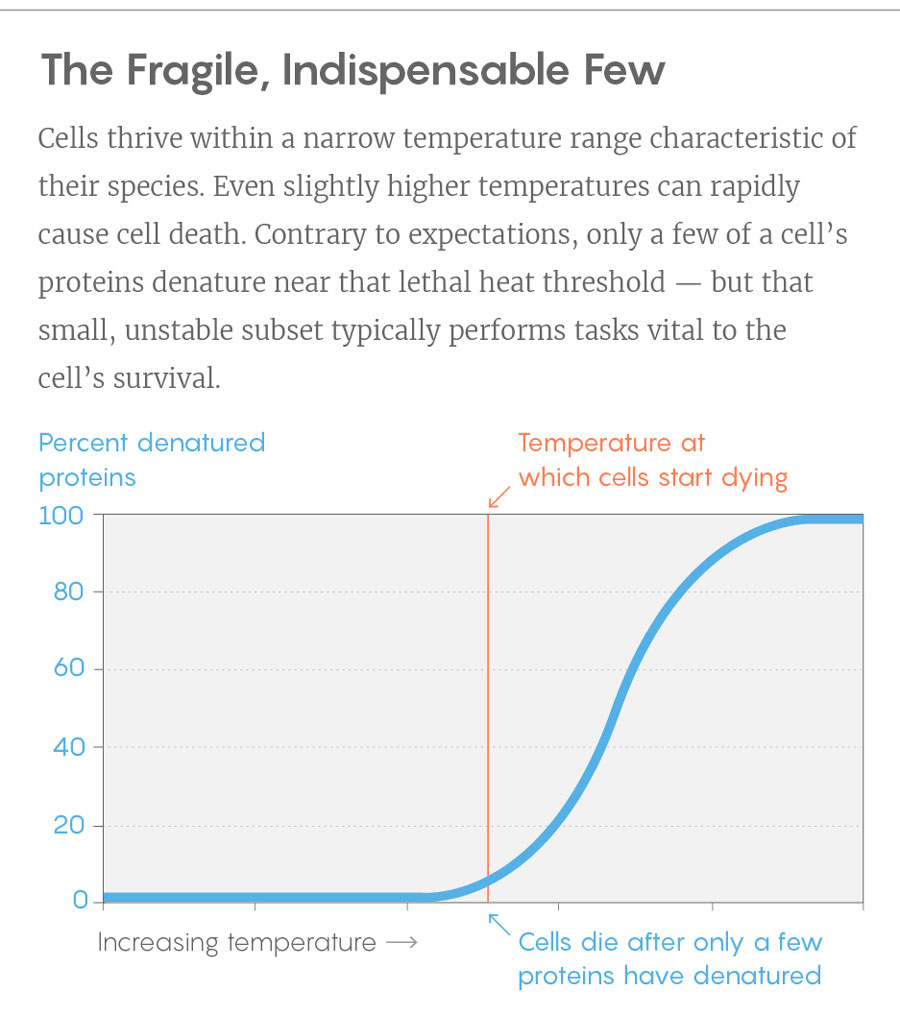
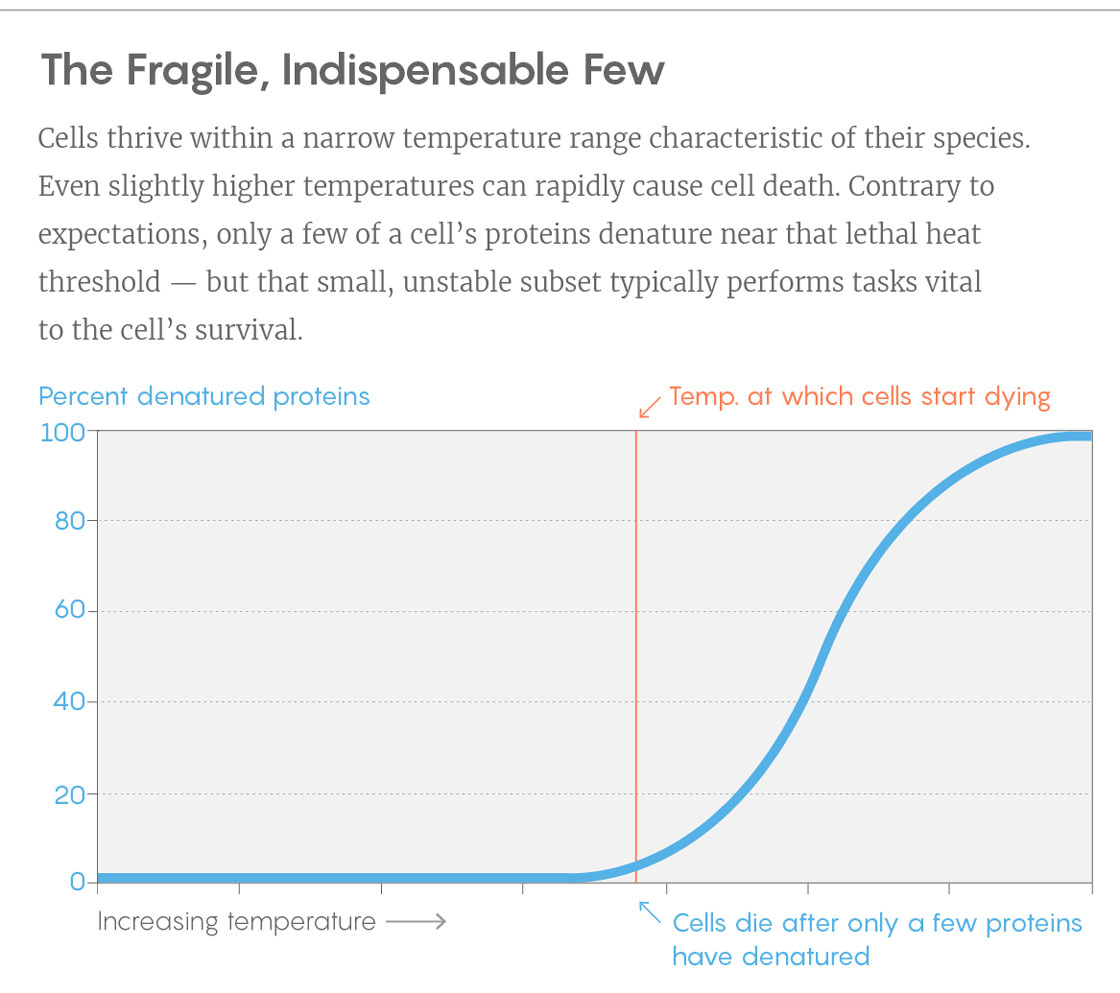
Lucy Reading-Ikkanda/Quanta Magazine
These findings could help researchers design proteins with stabilities carefully tuned to their needs. In many industrial processes that involve bacteria, for instance, raising the temperature increases yield — but before too long the bacteria die from the trauma of heat. It will be interesting to see if we can stabilize a bacterium by making those few proteins that disintegrate early more resistant to temperature, Picotti said.
Beyond all these observations, however, the group’s wealth of information about how easily each protein unfolds has some biologists especially excited. A protein’s stability is a direct measure of how likely it is to form aggregates: clumps of unfolded proteins that stick to each other. Aggregates, often a nightmare for the cell, can interfere with essential tasks. For instance, they are implicated in some serious neurological conditions, such as Alzheimer’s disease, in which plaques of denatured proteins gum up the brain.
But that doesn’t mean aggregation occurs only in individuals suffering from these conditions. On the contrary, investigators are realizing that it may be happening all the time, without obvious stressors, and that a healthy cell has ways of dealing with it. “I think this is increasingly recognized as a very common phenomenon,” said Michele Vendruscolo, a biochemist at the University of Cambridge. “Most proteins actually misfold and aggregate in the cellular environment. The most fundamental information obtained by Picotti is about the fraction of time in which any given protein is in its unfolded state. This fraction determines the degree to which it will aggregate.” Some proteins almost never unfold and aggregate, others do it only in certain situations, and still others do it constantly. The new paper’s detailed information will make it much easier to study why these differences exist and what they mean, he said. Some of the denaturing curves even show patterns that suggest the proteins were aggregating after they unfolded. “They’ve been able to monitor both steps — both the unfolding and the subsequent aggregations,” Vendruscolo said. “That’s the excitement of this study.”
While many scientists are interested in aggregates because of the damage they cause, some are thinking about the phenomenon from another angle. Drummond said it has become clear that some aggregates are not just wads of trash floating around the cell; rather, they contain active proteins that continue to do their jobs.
Imagine that from a distance, you see smoke billowing out of a building, he said. All around it are forms that you take to be bodies, dragged from the wreckage. But if you get closer, you may find that they’re actually living people, who escaped from the burning building and are waiting for the emergency to pass. That’s what’s happening in the study of aggregates, Drummond said: Researchers are finding that instead of being casualties, proteins in aggregates may sometimes be survivors. “In fact, there is a whole field that is now exploding,” he said.
Rather than being just a sign of damage, the clumping may serve as a way for proteins to preserve their function when the going gets tough. It might help protect them from the surrounding environment, for instance. And when conditions improve, the proteins could leave the aggregates and refold themselves. “They have temperature-sensitive [shape] changes that, if you don’t look too closely, look like misfolding,” Drummond said. “But there’s something else going on.” In a 2015 Cell paper, he and collaborators identified 177 yeast proteins that seem to regain function after being cloistered in aggregates. In a paper that appeared this past March, his team found that altering one of these proteins so that it couldn’t aggregate actually caused serious problems for the cell.
All in all, this work suggests that proteins are curiously dynamic structures. At first they might look like rigid machines, at work on fixed tasks for which one specific shape suits them. But in fact, proteins may morph into several different forms in the course of their normal duty. And in times of need, their shapes may alter so radically that they look as though they are expiring, when they are really fortifying themselves. At the molecular level, life may consist of constantly coming together and falling apart.
This article was reprinted on TheAtlantic.com.