Life’s First Molecule Was Protein, Not RNA, New Model Suggests

Scientists are using a computational model to envision how random chemical reactions on early Earth could have formed longer molecular chains capable of self-replicating and initiating the path toward cellular life.
Rachel Suggs for Quanta Magazine
Introduction
Proteins have generally taken a back seat to RNA molecules in scientists’ speculations about how life on Earth started. Yet a new computational model that describes how early biopolymers could have grown long enough to fold into useful shapes may change that. If it holds up, the model, which is now guiding laboratory experiments for confirmation, could re-establish the reputation of proteins as the original self-replicating biomolecule.
For scientists studying the origin of life, one of the greatest chicken-or-the-egg questions is: Which came first — proteins or nucleic acids like DNA and RNA? Four billion years ago or so, basic chemical building blocks gave rise to longer polymers that had a capacity to self-replicate and to perform functions essential to life: namely, storing information and catalyzing chemical reactions. For most of life’s history, nucleic acids have handled the former job and proteins the latter one. Yet DNA and RNA carry the instructions for making proteins, and proteins extract and copy those instructions as DNA or RNA. Which one could have originally handled both jobs on its own?
For decades, the favored candidate has been RNA — particularly since the discovery in the 1980s that RNA can also fold up and catalyze reactions, much as proteins do. Later theoretical and experimental evidence further bolstered the “RNA world” hypothesis that life emerged out of RNA that could catalyze the formation of more RNA.
But RNA is also incredibly complex and sensitive, and some experts are skeptical that it could have arisen spontaneously under the harsh conditions of the prebiotic world. Moreover, both RNA molecules and proteins must take the form of long, folded chains to do their catalytic work, and the early environment would seemingly have prevented strings of either nucleic acids or amino acids from getting long enough.
Ken Dill, a biophysicist at Stony Brook University, has been studying protein folding for decades. He’s now using that work to examine the chemistry-to-biology transition that took place four billion years ago.
Stony Brook University
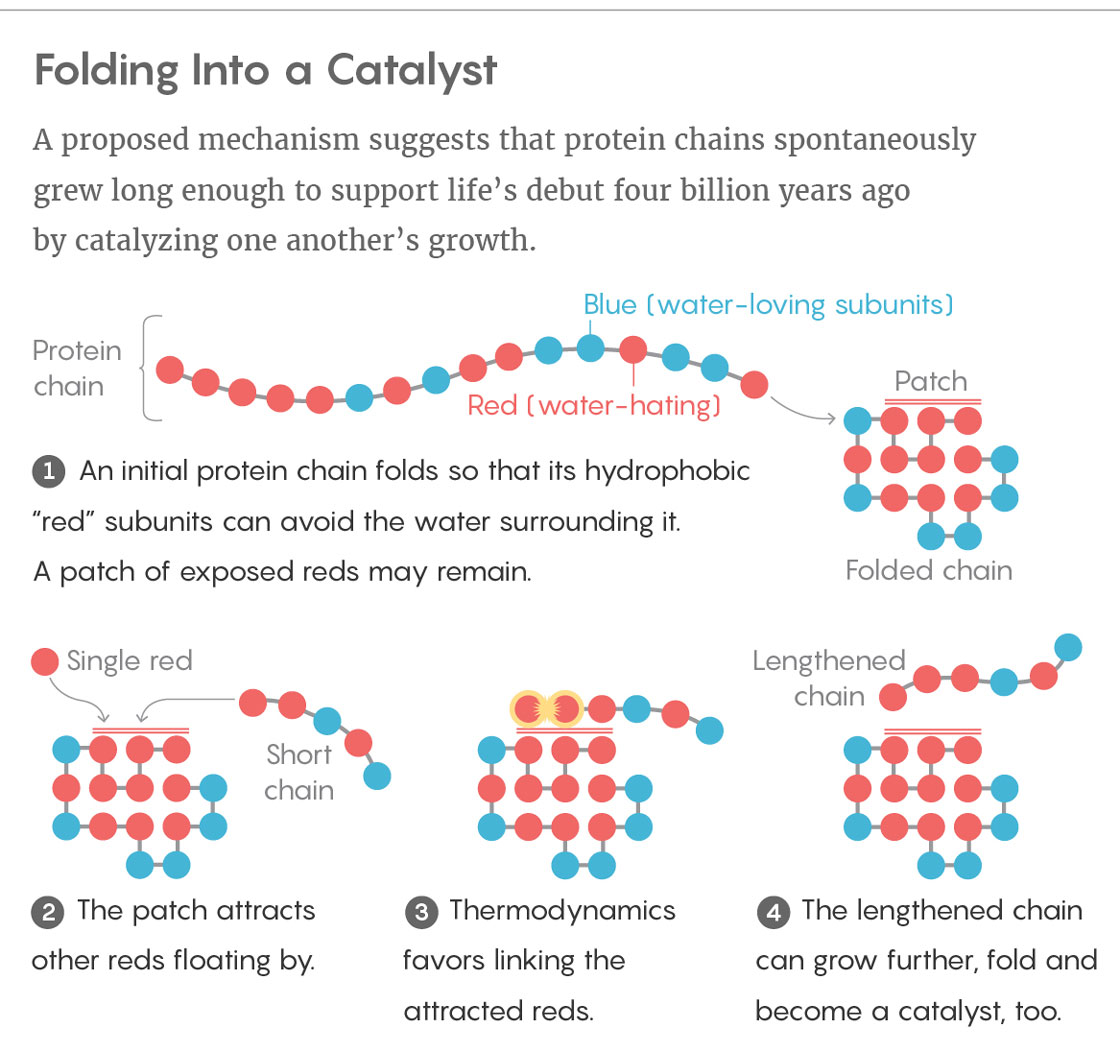
Ken Dill and Elizaveta Guseva of Stony Brook University in New York, together with Ronald Zuckermann of the Lawrence Berkeley National Laboratory in California, presented a possible solution to the conundrum in the Proceedings of the National Academy of Sciences (PNAS) this summer. As models go, theirs is very simple. Dill developed it in 1985 to help tackle the “protein-folding problem,” which concerns how the sequence of amino acids in a protein dictates its folded structure. His hydrophobic-polar (HP) protein-folding model treats the 20 amino acids as just two types of subunit, which he likened to different colored beads on a necklace: blue, water-loving beads (polar monomers) and red, water-hating ones (nonpolar monomers). The model can fold a chain of these beads in sequential order along the vertices of a two-dimensional lattice, much like placing them on contiguous squares of a checkerboard. Which square a given bead ends up occupying depends on the tendency for the red, hydrophobic beads to clump together so that they can better avoid water.
Dill, a biophysicist, used this kind of computation throughout the 1990s to answer questions about the energy landscapes and folding states of protein sequences. Only recently did he think of applying the model to early Earth — and to the transition from prebiotic chemistry to biology. “Chemistry is not self-serving, and biology is,” Dill said. “What were the first seeds of this self-servingness?”
The answer, he thinks, lies in foldable polymers, or foldamers. With his model, he generated one set of permutations of hydrophobic and polar monomers: the complete assortment of all possible red-and-blue necklaces up to 25 beads in length. Just 2.3 percent of these sequences collapse into compact foldamer structures. And just 12.7 percent of those — a mere 0.3 percent of the original set — fold into conformations that expose a hydrophobic patch of red beads on their surface.
This patch can serve as an attractive, sticky landing pad for hydrophobic sections of sequences floating by. If a single red bead and a red-tailed chain land on the hydrophobic patch at the same time, thermodynamics favors the two sequences linking together. In other words, the patch acts as a catalyst for elongating polymers, speeding up those reactions as much as tenfold. Although this rate enhancement is small, Dill said, it is significant.
Autocatalytic Origami
Most of those elongated polymers merely continue on their way. But a few end up folding, and some even have a hydrophobic patch of their own, just like the original catalyst. When this happens, the folded molecules with landing pads not only continue to form long polymers in greater and greater numbers, but they can also end up constituting what’s called an autocatalytic set, in which foldamers either directly or indirectly catalyze the formation of copies of themselves. Sometimes two or more foldamers can engage in mutual catalysis, by enhancing reactions that form one another. Although such sets are rare, the number of these molecules would grow exponentially and eventually take over the prebiotic soup. “It’s like lighting a match and setting a forest fire,” Dill said.
“That’s the whole magic of it,” he added, “the ability of a small event to leverage itself to much bigger events.”
Lucy Reading-Ikkanda/Quanta Magazine
And all that’s needed to spark this process are particular sequences of hydrophobic and polar components, which his model can predict. “Dill’s model shows you need only those two properties,” said Peter Schuster, a theoretical chemist and professor emeritus at the University of Vienna. “That’s a beautiful theoretical result.”
“It puts in doubt the vision of the origin of life that is based on the RNA world hypothesis,” said Andrew Pohorille, director of NASA’s Center for Computational Astrobiology and Fundamental Biology. To him and some other scientists, proteins seem like a “more natural starting point” because they are easier to make than nucleic acids. Pohorille posits that the information storage system found in the earliest rudiments of life would have been less advanced than the nucleic acid-based system in modern cells.
“People didn’t like the protein-first hypothesis because we don’t know how to replicate proteins,” he added. “This is an attempt to show that even though you cannot really replicate proteins the same way you can replicate RNA, you can still build and evolve a world without that kind of precise information storage.”
This fertile information-rich environment might then have become more welcoming for the emergence of RNA. Since RNA would have been better at autocatalysis, it would have been favored by natural selection in the long run. “If you begin with a simpler model [like Dill’s], something like RNA could appear later, and it would become a winner in the production game,” said Doron Lancet, a genomics researcher who has worked on his own simple chemistry-based model at the Weizmann Institute of Science in Israel.
Seeking Proof With Peptoids
Of course, the key to all this lies in actual experimentation. “Everything that goes back further than 2.5 to 3 billion years is speculation,” said Erich Bornberg-Bauer, a professor of molecular evolution at the Westfälische Wilhelms University of Münster in Germany. He described Dill’s work as “really a proof of concept.” The model still needs to be tested against other theoretical models and experimental research in the lab if it is truly to put up a good fight against the RNA world hypothesis. Otherwise, “it’s like the joke about physicists [assuming] cows are perfectly elastic spherical objects,” said Andrei Lupas, director of the department of protein evolution at the Max Planck Institute for Developmental Biology in Germany, who believes in an RNA-peptide world, in which the two coevolved. “Any significance ultimately comes from empirical approaches.”
That’s why Zuckermann, one of Dill’s co-authors on the PNAS paper, has begun working on a project that he hopes will confirm Dill’s hypothesis.
Twenty-five years ago, around the time that Dill proposed his HP protein-folding model, Zuckermann was developing a synthetic method to create artificial polymers called peptoids. He has used those nonbiological molecules to create protein-mimicking materials. Now he’s using peptoids to test the HP model’s predictions by examining how sequences fold and whether they would make good catalysts. In the course of this experiment, Zuckermann said, he and his colleagues will be testing thousands of sequences.
Ronald Zuckermann, a chemist at the Lawrence Berkeley National Laboratory, holds a model of one of the protein-like structures he’s formed using artificial polymers called peptoids. He’s now using peptoids to test the predictions of a new origin-of-life hypothesis. Berkeley Lab
That’s sure to be messy and difficult. Dill’s HP model is highly simplified and doesn’t account for many of the complicated molecular details and chemical interactions that characterize real life. “This means we will run into atomic-level realities that the model is not capable of seeing,” Zuckermann said.
One such reality might be that a pair of foldamers would aggregate instead of catalyzing each other’s production. Skeptics of Dill’s hypothesis worry that it would be far easier for the hydrophobic patches to interact with one another instead of with other polymer chains. But according to Pohorille, the potential for aggregation doesn’t automatically mean Dill is wrong about needing those hydrophobic patches to get autocatalysis started. “Modern enzymes aren’t just smooth balls. Enzymes contain crevices that assist the process of catalysis,” he explained. If there’s aggregation between the foldamers through their landing pads, it’s possible that the resulting structure could possess such features, too.
“Even if it seems unlikely, science has to consider all the hypotheses,” Bornberg-Bauer added. “That’s what Dill is doing.”
For now, at least, the RNA world hypothesis reigns supreme. Nevertheless, Dill and Zuckermann remain optimistic about what further research will yield. Dill plans to use the model to examine other questions about the origins of life, including how and why the genetic code arose. And Zuckermann hopes that the research — in addition to confirming (or refuting) Dill’s computations — will also help him make foldamers that can act as vehicles for drug delivery, synthetic antibodies or diagnostic tools.
“This model gives experimentalists like me a starting point,” Zuckermann said. “It lays down the challenge to find these primitive catalysts, to show how they work, to say: This could have really happened.”
This article was reprinted on ScientificAmerican.com.