Researchers Rethink the Ancestry of Complex Cells
Research into the origins of the complex cells called eukaryotes typically tries to trace lineages of the organisms back to a single ancestral cell. But now scientists are considering whether that common “ancestor” might really be an entire population of diverse cells.
DVDP for Quanta Magazine
Introduction
Our planet formed a little over 4.5 billion years ago, and if the most recent estimates are correct, it wasn’t long before life arose. Not much is known about how that happened because it’s maddeningly difficult to investigate. It’s also proved tough to study what happened next, during the first billions of years of evolution that followed, when the main domains of life emerged.
A particularly vexing mystery is the rise of the eukaryotes, cells with well-defined internal compartments, or organelles, which are present only in animals, plants, fungi and some microbes like protists — our evolutionary kin. The earliest eukaryotes left no clear fossils as clues, so researchers are forced to deduce what they were like by comparing the structural and molecular details of later ones and inferring their evolutionary relationships.
Right now is “an incredibly exciting time” for such research, said Michelle Leger, a postdoctoral fellow at the Institute of Evolutionary Biology in Barcelona, Spain. With modern genetic sequencing technologies, scientists can read the entire genomes of diverse life forms, and as microbial life is revealed in ever-increasing detail, new species and other taxonomic groups are coming to light. With that wealth of data, researchers are tracing lineages of organisms backward through time. “We’re trying to approach the problem from so many sides,” she said. “That’s pushing us closer to the first eukaryotes.”
This paramecium, a single-cell protozoan microbe, has a nucleus, mitochondria and other organelles that are hallmarks of eukaryotic cells.
Michael Abbey/Science Source
And those first eukaryotes may depart significantly from what most scientists expected, if some recent findings are any indication. Earlier this month, one team presented evidence that a signature event in eukaryote evolution — the development of the organelles called mitochondria — might have unfolded quite differently than was theorized. Meanwhile, other researchers have suggested that the earliest “ancestor” of all eukaryotes might not have been a single cell at all, but rather a mixed population of cells that avidly swapped DNA. The difference is subtle, but it might be important for understanding the evolution and diversity of the eukaryotes we see today.
The Ancestral Eukaryotes
The very first cells — the first life forms on this planet — were prokaryotes, but they were not all alike. Even early on, two very distinct lineages emerged, the archaea and the bacteria. The archaea might have been the first to thrive because even now they can survive in extreme environments like hot vents and super-saline pools. But it’s also possible that archaea and bacteria split from the first cells at the same time and began to diversify independently from the start. Figuring out definitively when and how the split occurred is probably impossible given how much time has passed; fossil evidence is nonexistent, and organisms from both branches have swapped genes extensively through horizontal gene transfer (as opposed to the “vertical” transfer of genes down through generations), which complicates analyses of their genomic history.
What we do know is that the story of eukaryotes began when some rogue archaeal cell split from the rest and founded what was long considered an entirely new domain of life. “First and fundamentally we are a very strange kind of archaea,” said Maureen O’Malley, a philosopher of biology affiliated with the University of Bordeaux and the University of Sydney.
It would be a struggle to distinguish the cells of this first eukaryotic common ancestor, or FECA, as such. It didn’t yet have a nucleus, for example. It didn’t have mitochondria to convert sugars and other molecules into more metabolically usable forms of energy. It didn’t even have microtubules, the structural proteins in eukaryotic cells that allow for compartmentalization by enabling the cell to shuttle things where they need to go.
No one really knows how eukaryotes came to possess those and other traits common to all eukaryotes but absent from other forms of life. But in a report in Nature Microbiology last week, a team of researchers from Europe and the U.S. offered a new theory about one of those milestones, the development of mitochondria. For decades, researchers have known that mitochondria are derived from bacteria that became internal symbionts of archaeal cells, but details of how that happened have been sketchy.
Anja Spang, a microbial ecologist at the Royal Netherlands Institute for Sea Research, Thijs Ettema, a microbiologist studying genome evolution at Uppsala University in Sweden, and their colleagues sought clues by looking at metabolic capabilities among the Asgard archaea, a superphylum that was discovered only a few years ago and is generally recognized as having the most in common with eukaryotes.
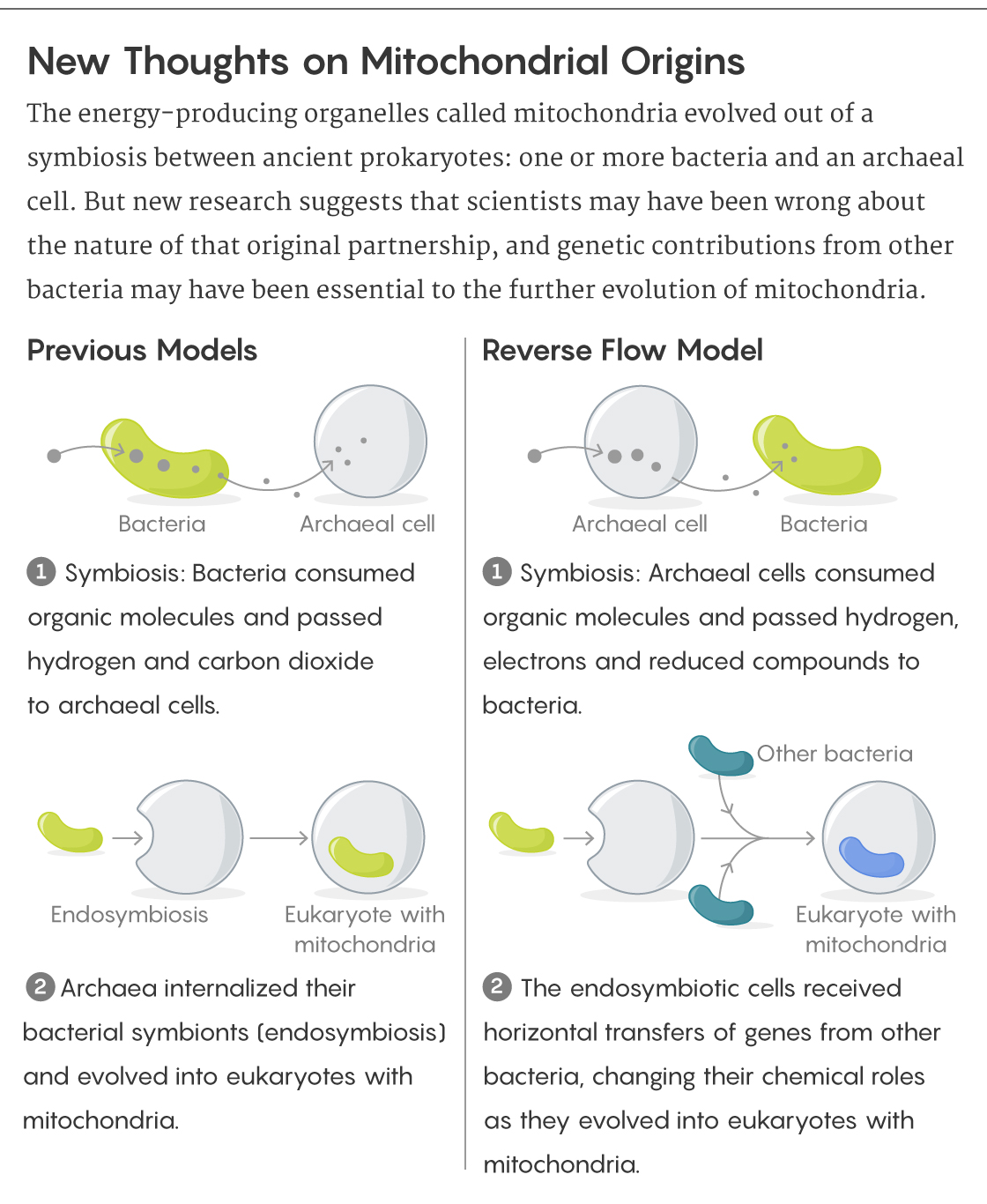
The scientists concluded that mitochondria most likely arose out of a partnership between archaeal cells that fermented certain small organic molecules and alphaproteobacteria that survived by oxidizing certain other ones: The bacteria could use the electrons and hydrogen that the archaeal cells shed as wastes. (The researchers call this the “reverse flow model” because according to a previously popular theory, the bacteria would have donated hydrogen to the archaea’s metabolism.)
Lucy Reading-Ikkanda/Quanta Magazine; adapted from DOI: 10.1038/s41559-019-0796-3
“Such associations could render growth on some small organic substrates more favorable,” Spang and Ettema explained in an email. For example, some modern archaea that live under oxygen-free conditions and metabolize hydrocarbons depend on bacteria to accept their electrons. “A similar type of interaction may have characterized the presumed archaeal ancestor of eukaryotes.”
Over time, horizontal transfers of genes from other bacteria would have provided more of the machinery for the metabolic processes performed by mitochondria as we know them. Meanwhile, gene transfers between archaeal hosts and their bacterial symbiotes, along with the loss of some superfluous genes on each side, would have cemented what had been separate symbiotic cells into a permanently unified eukaryotic state.
The researchers note that although this theory could explain the origin of the mitochondrion, it is silent on the origins of other important organelles. “Supposedly, we should start referring to a eukaryotic cell once the nucleus had evolved,” Spang and Ettema wrote. “At this point, it is still unclear whether this happened before or after the mitochondrial endosymbiosis.” They also note that if ancient archaea did start adopting some eukaryotic features before the symbiosis with alphaproteobacteria began, it might have helped the transition. Filaments of the protein actin, for example, could have stabilized contacts between the hosts and symbionts and improved the coupling of their metabolisms.
Overall, the genesis of eukaryotes remains mysterious because all eukaryotes alive today arose from an organism that was already complex. Somehow, over an unknown number of millennia, FECA turned into the last eukaryotic common ancestor, or LECA — an organism ancestral to every other subsequent eukaryote living or extinct, including ones currently unknown to science. LECA is a lot easier to imagine because it probably looked similar to some of today’s microbial eukaryotes. “It turns out that everything that has a nucleus also has mitochondria, a Golgi apparatus and everything else,” said W. Ford Doolittle, a molecular biologist at Dalhousie University in Nova Scotia. “LECA appears to have already been a fairly sophisticated eukaryotic cell.”
In fact, LECA is seemingly so straightforward that some say it’s downright boring. “The one thing that nobody really bothers to argue about at all is the nature of the last eukaryotic common ancestor,” said Anthony Poole, a molecular evolutionist at the University of Auckland in New Zealand.
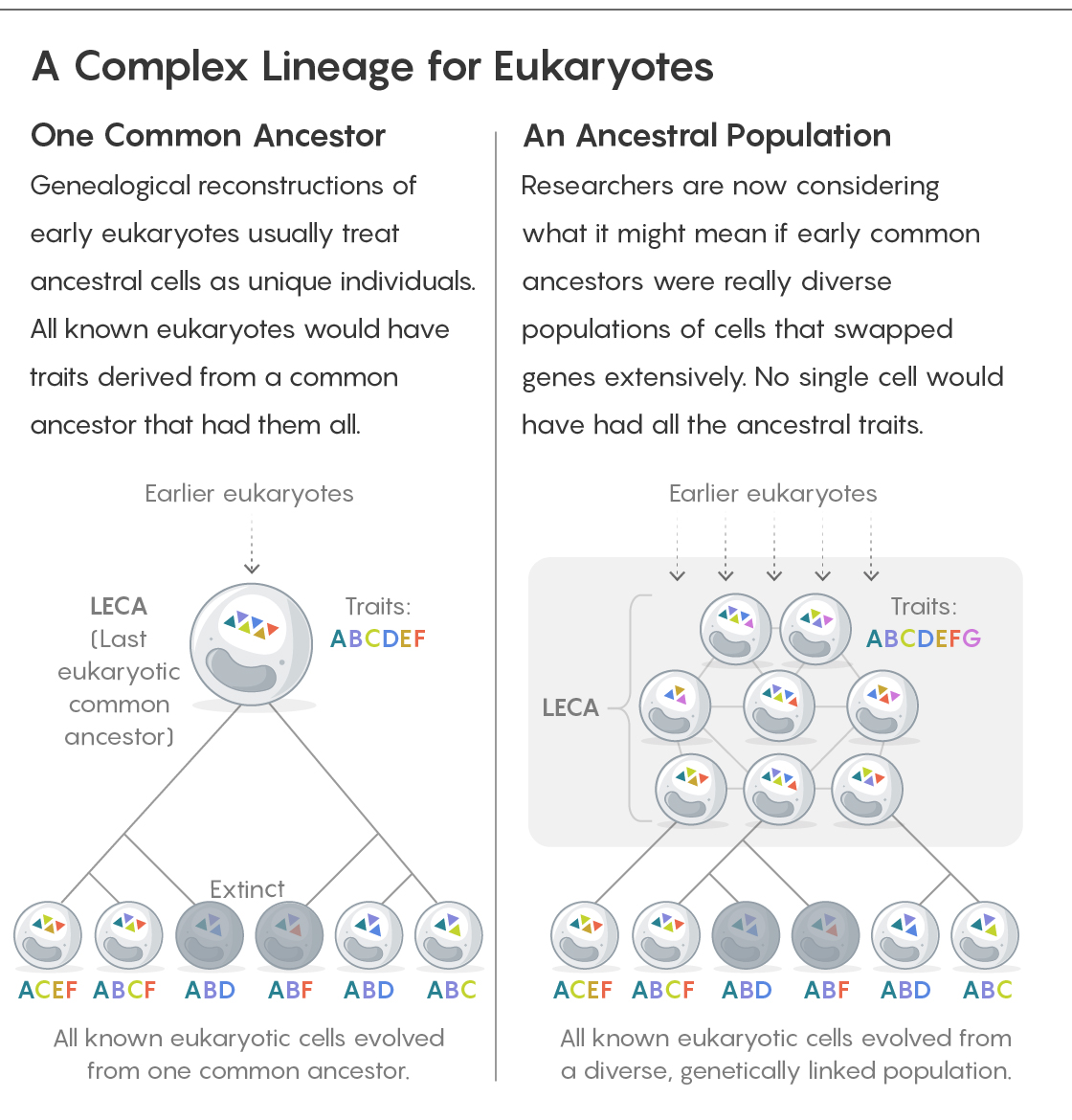
Except, some are bothering to argue about it — because LECA is usually discussed as one cell, the singular ancestor of all eukaryotes. To O’Malley, that’s wrong. “Obviously LECA can’t have been a single cell,” she said. That error, she thinks, comes from people thinking about the genealogy too simplistically and confusing ancestry with ancestors. “Genealogical thinking just picks out that lineage with all its divisions back to that single-cell lineage.”
In an essay for Nature Ecology & Evolution, O’Malley and her colleagues discussed the implications if LECA was not a single cell but really a population of genetically diverse cells, none of which had all the characteristics associated with eukaryotes today. “When we’re talking about LECA, we’re probably talking about an ancestral state, a genomic state that we don’t know was one single cell,” O’Malley said.
“What we really wanted to do with this paper was just generally start a conversation between people working on reconstructing the last eukaryotic common ancestor to think about how they conceive of LECA, and whether they could imagine that the genetic variation in a hypothetical population might explain some of the patterns that they see,” explained Leger, who is a co-author on the paper.
O’Malley, Leger and their colleagues argue that to truly understand LECA and decode its genomes — and to get a more complete picture of what all eukaryotes are — we need to understand what that ancient population was like.
One Cell or Many?
Bill Wickstead of the University of Nottingham and his colleagues are among those trying to reconstruct LECA. Their effort centers on building a proteome — the complete collection of proteins that LECA was probably capable of making. This is done by taking genomes and proteomes from diverse eukaryotic lineages and using statistics to determine which traits are most likely to have been present in their common ancestor, and which arose as independent evolutionary innovations or were passed horizontally among lineages. Molecular biology like this offers the best hope for revealing LECA.
But a key point about this approach, Wickstead says, is that it doesn’t strictly matter whether the ancestral proteome and genome it deduces is in a single cell or if it’s distributed across a population of them. It’s a statistical extrapolation of genomic data that doesn’t trace back cellular divisions.
“From the point of view of the LECA genome … it really makes no difference whether LECA was an individual cell or a collection of cells” that exchanged genes in addition to replicating, Wickstead said.
But, as O’Malley and Wickstead both point out, this distinction is not just a matter of semantics. Whether the genome that gets reconstructed from current data was in one cell or spread across many of them is vital to understanding how that genome was used — basically, it’s the difference between genetics and cell biology, Wickstead says.
He and his colleagues are mostly interested in reconstructing LECA’s genome and proteome to understand what biological abilities it had. “But it does make a difference, really, to think about whether or not they could all exist within an individual cell, or if you would have to then break them out, because there would be conflicts within that,” he said.
Leger agrees that knowing whether LECA was one cell or many would help researchers make better sense of the genomic data. “If you try and look at all of the features that are commonly shared by many eukaryotes today, and you try to reconstruct which features should have been present in the last eukaryotic common ancestor going from just those features, you end up with the last eukaryotic common ancestor cell that has an impossibly large genome encoding far too many proteins to be normal,” she said.
A similar problem arises with a more familiar organism, the common bacterium Escherichia coli, which is a single species divided into many genetic strains. If you take the genomes from multiple strains of E. coli, it’s clear that different strains have different genes — not just different variants of genes, but entire gene families that are present or absent in various cell lines. Each individual bacterium has a genome with roughly 4,200 to 5,600 genes. Somewhere between 2,200 and 3,100 of those are found in all E. coli; the rest are drawn from a total pool of at least 89,000 possible “accessory” genes. And although they’re not central to the core existence of the bacteria, they influence its survival in many ways. The variation in the thousands of accessory genes explains why some strains are virulent while others are harmless, and how some can survive in certain habitats or on food sources that others can’t.
Accessory genes can be horizontally passed from one strain to another, so if we want to understand the total capabilities of E. coli as an organism, we need a complete picture of the genomic variation in the species, or what researchers call a pangenome.
Lucy Reading-Ikkanda/Quanta Magazine
The concept of a pangenome arose in the early 2000s, when scientists realized that reference genome sequences of pathogenic bacteria — the digital databases compiled as standardized descriptions of their genomes — failed to capture the total genetic variation of the organisms. Since then, scientists have realized that pangenomes also play an important role in prokaryotic life. But since the sharing of genes through horizontal gene transfer is much less common in eukaryotes, it’s long been assumed that pangenomes have only limited relevance to understanding eukaryotic species.
That view is slowly changing. A recent analysis of genomes from four medically important pathogenic fungal species found that they, too, have pangenomes: 10-20 percent of their genome is comprised of accessory genes responsible for important traits like resistance to antimicrobial compounds.
Even our species has a pangenome. “When we first sequenced the human genome, it was hailed as fantastic — ‘We now have the blueprint for all humans!’ But of course we didn’t,” Wickstead said. A recent study found that nearly 10 percent of the genes from 910 people of African descent weren’t in the human reference genome, for example. “And the missing content of genes within individual genomes is part of the diversity of humans, hence a response to drugs, and the environment, and all sorts of very important things,” Wickstead said.
If extant eukaryotes have pangenomes, and extant prokaryotes do too, it would be odd to think that early eukaryotes didn’t. The “genome” that Wickstead and other scientists reconstruct when they try to deduce what LECA looked like is probably that pangenome.
That all makes sense to J. Peter Gogarten, an evolutionary biologist at the University of Connecticut. To him, the paper by O’Malley, Leger and colleagues crystallizes the idea that “to understand the origin of eukaryotes, we might need to move beyond reconstructing the tree of cells” and instead “focus on the network that describes the genome’s evolutionary history.” That’s something he’s been advocating for a while, he says. He thinks that moving away from looking at LECA as a single cell and recognizing it as a population of different cells might help us peer even further back into evolutionary history to the mysterious first eukaryotes.
Still, even Gogarten sets limits to that speculation: LECA may have been a population of cells, but he says he’s not convinced it was a large population with a vast and diverse pangenome. Larger populations, larger pangenomes or both likely came into play during the transition between FECA and LECA, which is why viewing these ancestors as populations rather than as individuals may help to illuminate the origins of eukaryotes. But Gogarten thinks that by the time LECA existed, things had become more settled.
Poole agrees. Not many of the features that most experts would consider to be critical for LECA, he said, are compatible with a pangenome explanation for their diversity. “We don’t have a model where we say, ‘You’re sharing half a ribosome each across two cells.’ Because that’s just physically implausible.”
But the pangenome might become more relevant as scientists gain a greater understanding of the diversity in behaviors among those early cells and in the environments they inhabited. That’s part of why Leger thinks it’s worth considering whether LECA might have had a big, diverse pangenome. Key aspects of metabolism, such as the ability to process certain sugars, could have been distributed in the population rather than in every cell. And that might have allowed the organism to colonize more environments than other microbes.
If so, that could help explain why eukaryotes diversified so quickly, Leger says. A diverse LECA population spread across many different environments could have led to many semi-isolated subpopulations prone to interbreeding. That’s a scenario that fosters diversification, as seen when species colonize islands today.
Nevertheless, others are skeptical that the pangenome of LECA would have been that big. The pangenomes of eukaryotes today are small in comparison with those of prokaryotes, which makes some scientists doubt that LECA’s pangenome was any larger than, say, ours. “I would agree that species have pangenomes, and LECA was a species, and LECA had a pangenome,” Doolittle said. “But I don’t see any reason to suppose that LECA was a species any different than the other species that are on the tree. It just happens to be the deepest node.”
The Basis of All Evolutionary Reasoning
Despite the debate surrounding the extent of LECA’s pangenome, many of O’Malley and Leger’s colleagues do agree that it makes sense to think of LECA as a population of cells. But there has been some pushback to that idea, too. According to O’Malley, some scientists insist that LECA had to be a single cell, one that split and then split again and again and again, eventually giving rise to all other eukaryotic cells. “There’s something, to me, very curious about this deep attraction to the genealogical view,” she said.
To her, viewing LECA as a population is the only way to truly understand how it arose, and how it led to the diversity of eukaryotes alive today. “Populations are the basis of all evolutionary reasoning,” she said: By definition, evolution and natural selection act at the population level. “It’s not a matter of cells, it’s always a matter of populations.” That’s why she and her colleagues “push that a bit harder,” she said, to emphasize that they need to think about populations when reconstructing the history of eukaryotes “because this affects how you reason about the environment, how you reason about the genes.”
It’s especially important to the attempt to reconstruct LECA, because ultimately, the organism that existed then likely wasn’t just comprised of the traits seen in eukaryotes or their closest living relatives today. “I’m not sure if one can understand evolution if you only look at what gets retained,” she said. “We have to understand the population stage to understand why certain things got lost along the way.”
The trouble is, we may never know what LECA looked like because no fossils or remnants of DNA will ever reveal its nature directly. Even the best genomic methods can’t literally turn back time and allow us to watch how a sequence changed. It’s basically impossible to concretely determine what LECA’s genome or pangenome looked like.
But that doesn’t mean it’s not worth pondering. LECA is where we all come from — “the raw material from which the diversity of eukaryotes arose,” as Wickstead put it. And just because we can’t see a way to rigorously test competing hypotheses now doesn’t mean we won’t be able to in the future. “I think it’s obviously quite important to understand what was back then, and what kind of biology was going on, in order to try and understand how the lineages then evolved from that point,” he said.
Simply asking these kinds of questions reveals gaps in our understanding of the eukaryotes alive today, in Leger’s view. “There’s so much that we still have to learn about microbial eukaryotes in general and just how they behave, what’s normal for them,” she said.