Where Do New Genes Come From?
Introduction
The evolution of new genes often goes hand in hand with the emergence of novel traits in species as they evolve. One of the great riddles in evolutionary biology has therefore always been how genetic novelty arises.
For the half past century or more, most biologists agreed with the conclusions of the geneticist Susumu Ohno in his influential 1970 book Evolution by Gene Duplication. While acknowledging that the first genes had to come from somewhere, he wrote: “Yet, in a strict sense, nothing in evolution is created de novo. Each new gene must have arisen from an existing gene…”
This explanation seemed sound because truly de novo genes would have to emerge through evolution acting on the abundant “nongenic” DNA (often dismissed as junk) between genes. It was hard to imagine how that could happen. Cells’ fitness generally depends on the smooth functioning of networks of genes that have coevolved to work together over millions of years. Genes derived from other genes have a better chance of blending into those networks. In comparison, the fairly random transcripts from nascent de novo genes seem as though they should be, at best, inconsequential and more likely harmful to cells’ prospects. “The received wisdom is that random sequences are more likely to mess things up than to make them better,” said Aoife McLysaght, a geneticist at Trinity College Dublin.
But in the past 15 years, evidence for de novo genes has steadily accumulated, so much so that the debate has shifted from whether de novo genes exist to how much they contribute to evolution and adaptation.
Recent experiments by McLysaght and other researchers have begun to quantify how often de novo genes occur in a wide range of organisms. Their estimates vary but the answers suggest that for many genes that are known to be young or novel, the de novo mechanism seems to be at least roughly on par with the alternative Ohno described — and sometimes even more common.
De novo genes “represent a really unprecedented or unrivaled kind of genetic novelty,” said Caroline Weisman, a doctoral student in biophysics at Harvard University who is conducting research into the origin of genes. “That’s a really exciting possibility for evolutionary biologists who are thinking about how things like novelty evolve.”
Many Ways to Become an Orphan
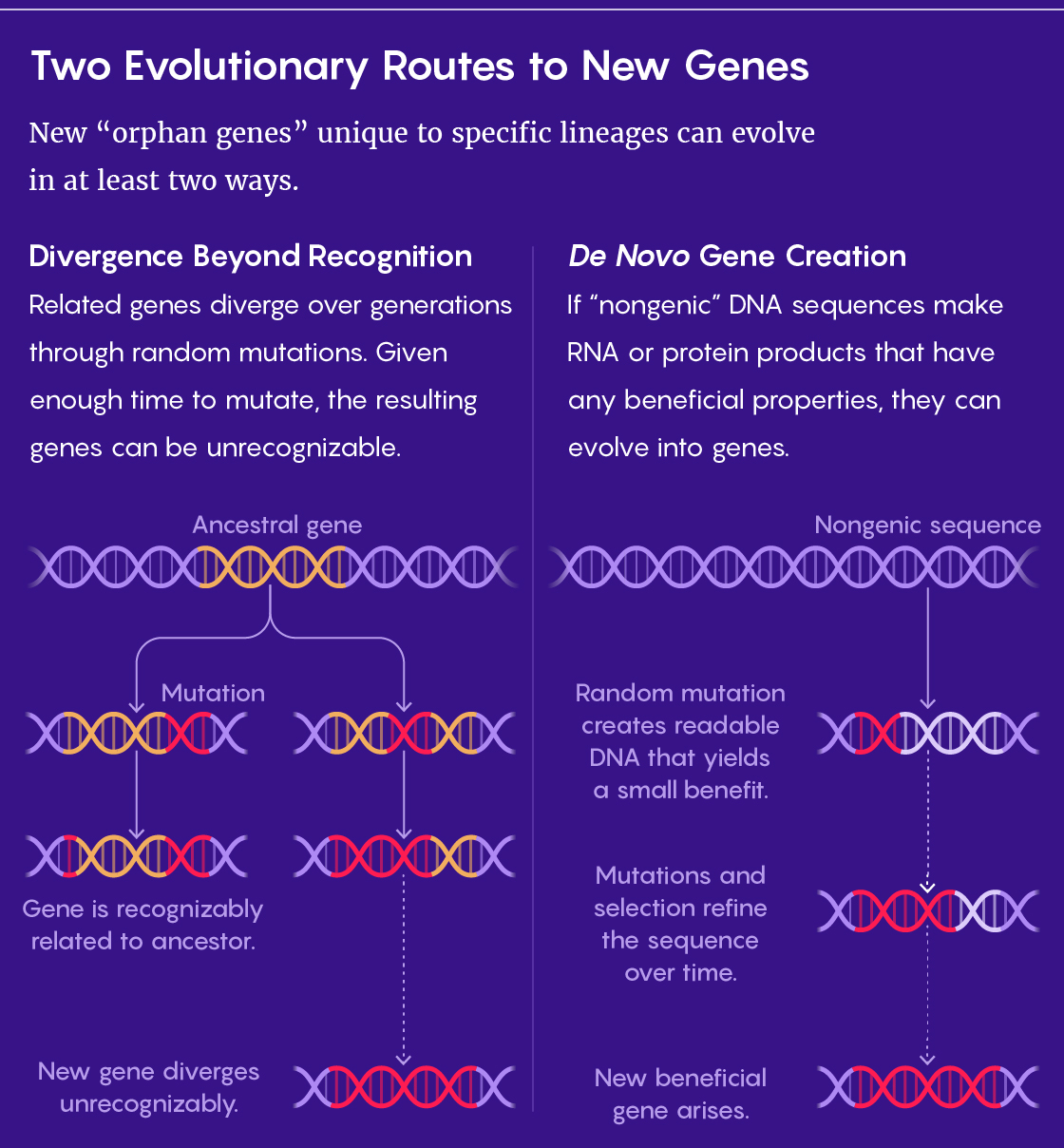
Most of the genes in every species can also be found in at least one other species. The genes may have slightly different sequences in each instance, but they look enough alike to be recognizably related — which they typically are through evolution. Random mutations make the sequences diverge over time, but homologous genes (or homologs) can still be sorted into families by their similarities. For example, the genes for all the slightly different hemoglobin molecules found in humans and other mammals belong to one family.
Ohno introduced the theory that a divergence mechanism could explain how genes with new functions arose. In his work, he showed that new genes could be born through the duplication of older ones, followed by mutations that made the two homologs diverge in function as well as sequence.
Yet as whole genomes became more available and researchers scoured them for information, it seemed that pieces were missing from the puzzle. Some genes did not seem to belong to any family. These “orphan genes” appeared specifically in certain lineages and had no obvious ancestors or cousins. The question then focused on how these orphan genes came to be.
Lucy Reading-Ikkanda/Quanta Magazine
The default assumption was that it was Ohno’s mechanism taken to an extreme — divergence beyond recognition. Orphan gene sequences could have evolved so quickly, or for such a long time, that they lost their family’s resemblance.
Other explanations were possible but, according to McLysaght, they seemed less likely. Orphan genes could enter a lineage through the horizontal transfer of whole or partial genes from bacteria or viruses, for example, but few of the identified orphans in complex organisms seemed as if they could have come from bacteria. Theoretically, a gene could also be orphaned if all of its homologs in other lineages were coincidentally lost through evolution — but that too seemed improbable to be a routine explanation. And then there was the de novo possibility, but that came with its own hurdles.
Still, researchers kept finding orphan genes that looked convincingly as if they had evolved de novo. In 2006 and 2007, for example, the geneticist David Begun at the University of California, Davis, identified genes in the testes of fruit flies that had evolved from nongenic sequences. Gradually, the question shifted from whether de novo genes existed to how common they were.
During the past decade, researchers have vigorously argued about the relative importance of de novo gene creation and divergence beyond recognition. But there was still no easy way to look at orphan genes and determine how they arose. “The field was hamstrung by that, in a sense, because if you can’t really know how many are real [de novo genes], and what’s the significance of this phenomenon, then you’re a bit stuck,” McLysaght said.
Location, Location, Location
To bring some clarity to that debate, McLysaght and her former postdoctoral fellow Nikolaos Vakirlis (now at the Alexander Fleming Biomedical Sciences Research Center in Greece), along with their collaborator Anne-Ruxandra Carvunis at the University of Pittsburgh, set out to quantify what proportion of the orphan genes in flies, yeast and humans could be explained by sequence divergence.
They took a novel approach to that analysis, as they described in a paper in eLife in February. Scientists usually check whether genes are homologous by comparing their nucleotide sequences (or the amino acid sequences of the proteins they encode). McLysaght’s team looked instead at each gene’s position relative to its neighbors — a property that geneticists call the gene’s synteny.
McLysaght offered this analogy to explain their approach: Suppose you start with an ordered deck of playing cards and lightly shuffle them. The first two cards off the top of the deck are the 9 and 10 of clubs; you keep the third card face down; the fourth and fifth cards are the queen and king of clubs. You could guess with reasonable confidence that the hidden card is the jack of clubs because the odds are better that the complete sequence survived than that the middle card alone was disturbed.
Similarly, the order of neighboring genes on a chromosome is mostly conserved through evolution. Pieces of chromosomes get resorted significantly, but within those shuffled blocks, the arrangement of genes tends to stay intact. The researchers made a conservative assumption that if a gene’s neighbors appear in the same order in another species, then the gene is likely to correspond to whatever is sandwiched between them in the other species as well — even if the sequences don’t match.
Using the synteny method, the researchers estimated that at most a third of orphan genes in flies, yeast and humans could be explained by divergence beyond recognition. “The rest must be explained by other ways, and the de novo origin is the best way to explain those,” McLysaght said.
Rates of Divergence
Weisman and her Harvard advisers Andrew Murray and Sean Eddy used a slightly different method to address the same problem in work they described recently in a preprint on the biorxiv.org server and have submitted to a journal for peer review. “The whole question here is, if I can’t detect a homolog outside of some organism or some group, is that because the homolog is there and I can’t detect it, or because the homolog isn’t there?” Weisman said.
To find out, she looked at a group of related yeast species and Drosophila fruit fly species and estimated the rates at which mutations accumulated within their gene families. She could then determine statistically whether the homolog for a gene in one species would even be detectable in distantly related species. That allowed her to identify cases where “your result that the gene looks like an orphan is totally explainable just through the gene evolving normally and your search software not being omniscient,” she explained.
Weisman estimated that somewhere between 55% and 73% percent of the orphan genes in these yeasts — a majority — were explained by divergence; that figure is higher than McLysaght’s synteny approach suggested. Nevertheless, to Weisman, it’s reassuring that her method and McLysaght’s fundamentally different one converged on the conclusion “that there is some decidedly nontrivial number of these genes that probably are just due to divergence.” She added, “Even if it’s 30% or 50% or 80%, either way you slice it, it’s clearly a problem for people who want to study [de novo genes] by studying orphan genes.”
Li Zhao, a geneticist at Rockefeller University who was not involved with either Weisman’s or McLysaght’s work, agrees that both papers reach roughly the same conclusion about the origins of orphan genes, although one emphasizes the abundance of de novo genes and the other the abundance of ones from divergence. “One paper is talking about this glass being half full, and the other is describing it as half empty,” she said.
Given that mixture of origins for orphan genes, Zhao continued to say, a good way to study the de novo ones might be to focus on the very young ones. If a de novo gene has originated recently, it should still be possible to identify the corresponding nongenic sequence in other species from which it evolved, she explained. That would serve as proof that the orphan gene is truly de novo.
How Function Emerges
A good illustration of this is a 2019 study of young de novo genes in wild Asian rice (Oryza) led by Manyuan Long, a geneticist at the University of Chicago who has pioneered research into novel genes since the early 1990s. Long and his colleagues identified about 175 genes that originated de novo within the last 3.4 million years; they could tell that these genes were de novo because corresponding nongenic sequences were still recognizable in closely related species. These de novo genes appeared to be biologically active — that is, they were transcribed into RNA and translated into peptide chains, and most of them showed signs of being shaped by natural selection.

In wild Oryza rice, researchers led by Manyuan Long of the University of Chicago identified 175 de novo genes that evolved within the past 3.4 million years.
Courtesy of Manyuan Long Lab, with additional efforts of Shengqian Xia, Tao Yang and Yidan Ouyang
Long’s study confirmed that de novo genes were relatively abundant and functionally important. But it left open the question of exactly how a nongenic sequence could become a functional gene. One possible answer is the “proto-gene” hypothesis put forward by Carvunis and her colleagues in a 2012 Nature paper: Incipient genes could begin as stretches of DNA that get made into RNA and protein products that don’t initially do anything. Under the right environmental conditions, though, these proto-genes could provide some advantages, and thus start evolving under selection.
Carvunis, Vakirlis, McLysaght and their colleagues tested that idea experimentally in a Nature Communications paper that appeared in February. First, they computationally identified DNA sequences in yeast that seemed to fit the definition of proto-genes by being evolutionarily young and actively transcribed but not making functional proteins. Then they saw what happened to the fitness of the yeast when these sequences were either deleted or overexpressed.
Deleting these proto-gene sequences didn’t seem to be harmful; that made sense because they weren’t contributing to the yeast’s well-being. But to the researchers’ surprise, when about 10% of the proto-gene sequences were overexpressed, they enhanced the yeast colonies’ growth. In fact, overexpressing these proto-gene sequences was more often beneficial than overexpressing established functional genes (evolution has presumably already set an optimal level of expression for them). “We didn’t necessarily expect that these somewhat random sequences would have this potential to add to the fitness,” McLysaght said.
According to Vakirlis, those results suggest that the proto-genes have high adaptive potential: Their effects may not be well defined, but they can potentially contribute to the cell in many ways. That potential is what evolution can explore over time if it refines the sequences into functioning genes.
“We show that emerging sequences can be adaptive,” Carvunis said.
The researchers also observed that the beneficial proto-gene sequences had something in common: Protein products translated from them would generally have domains that might enable them to perch in the membrane of a cell or organelle. The researchers are now investigating how, by situating itself there, a protein might increase its chances of doing something significant for a cell.
Although their study demonstrated the adaptive potential of emerging de novo genes, the actual contribution of de novo genes to adaptation might always “remain somewhat cloaked in mystery,” McLysaght said. As mutations accumulate in de novo genes, it gets harder to identify the nongenic sequences from which they came. Past some uncertain deadline, it may always be impossible to prove that an old gene arose de novo. Pinning down the true number of de novo genes and their contribution to novel adaptations in most complex organisms may therefore be an intractable problem.
Still, Long emphasized that orphan genes have biology worth investigating regardless of their origin. Weisman thinks that may be particularly true of genes whose divergence seems to have suddenly accelerated at some recent point in their evolution: They might be able to tell us about how novel biological functions evolve.
For the creation of orphan genes, “we know there’s a diversity of mechanisms,” Begun said. But “the guiding principles for why certain biological processes might have more de novo gene evolution, while others might have more duplication and divergence — that, we don’t really have a grip on yet.”
Vakirlis agreed about how many questions still needed to be addressed. “I don’t think in this field there’s anything that is well established yet other than the fact that de novo genes are real and they do appear to be widespread — and how widespread they are will vary depending on who you ask. It’s a very dynamic situation where we learn more and more by the year,” he said.